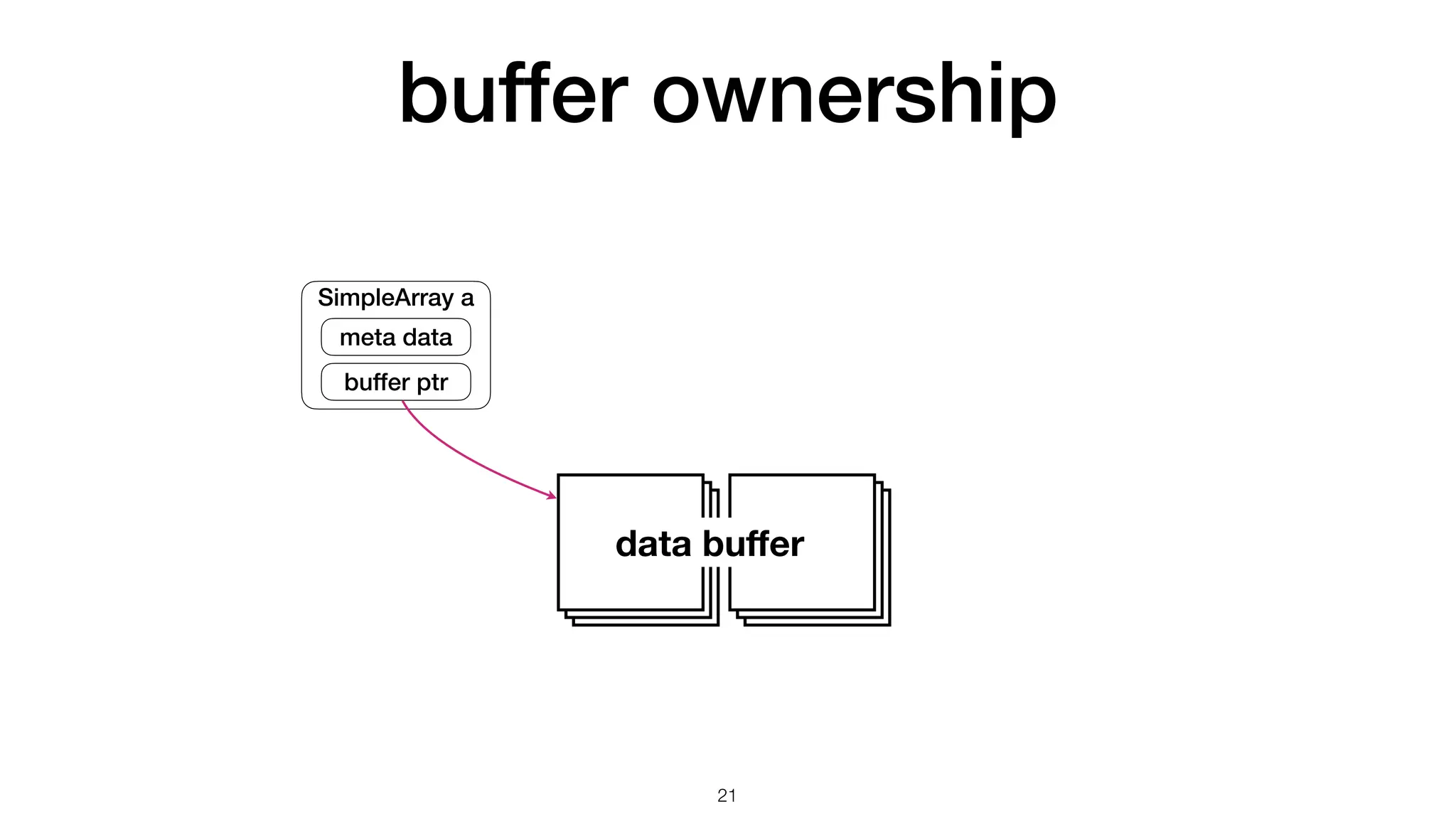
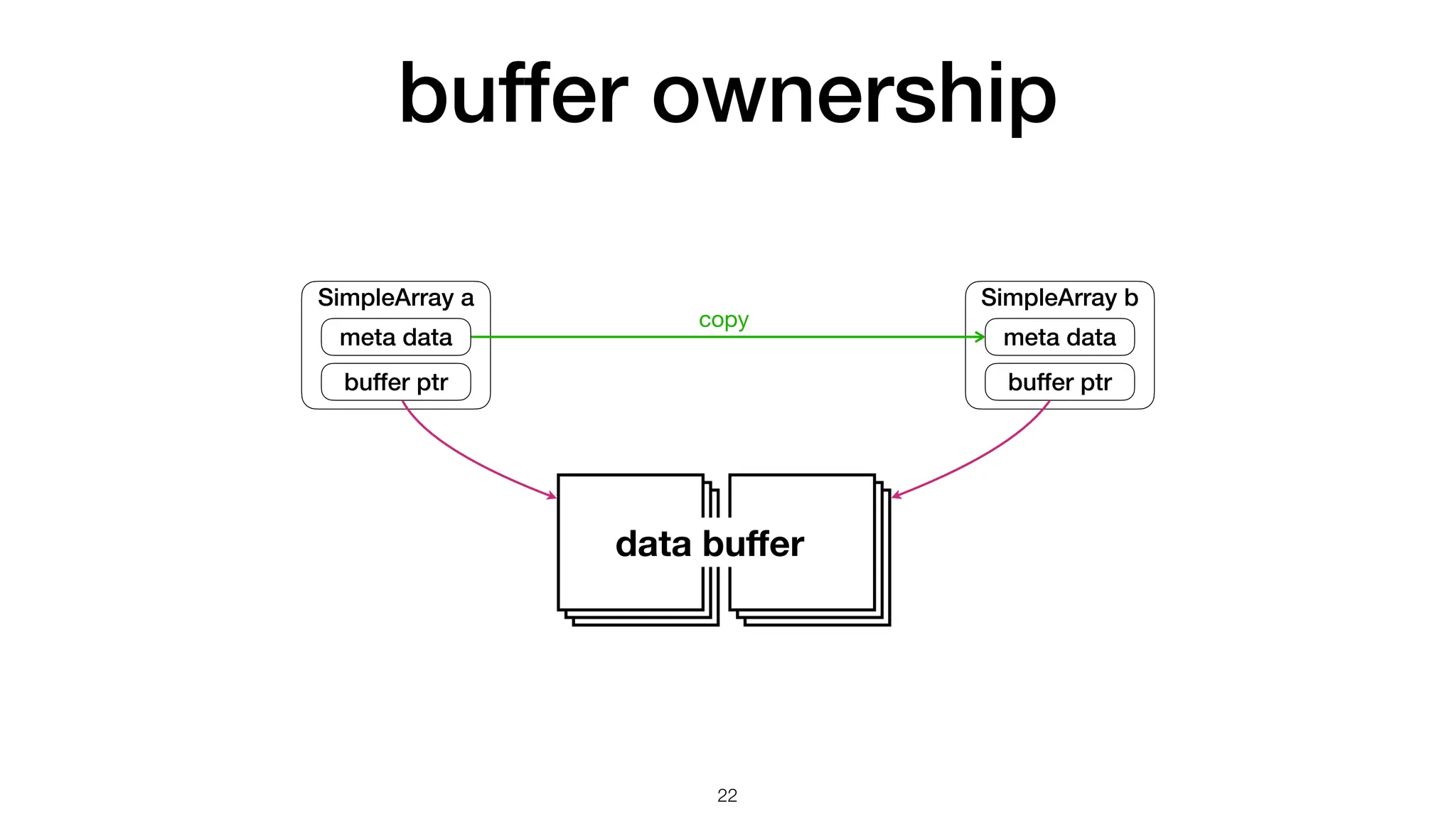
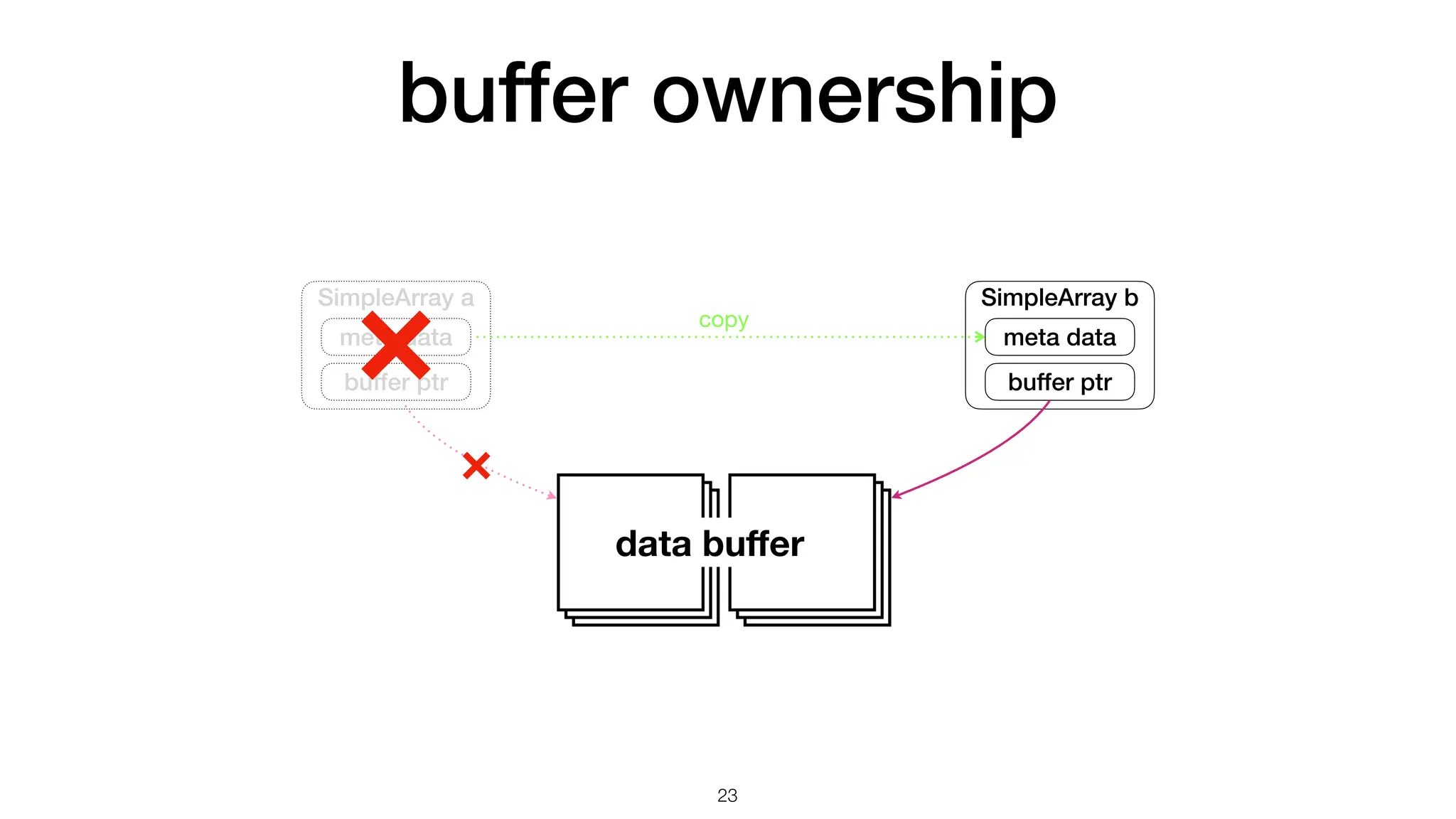
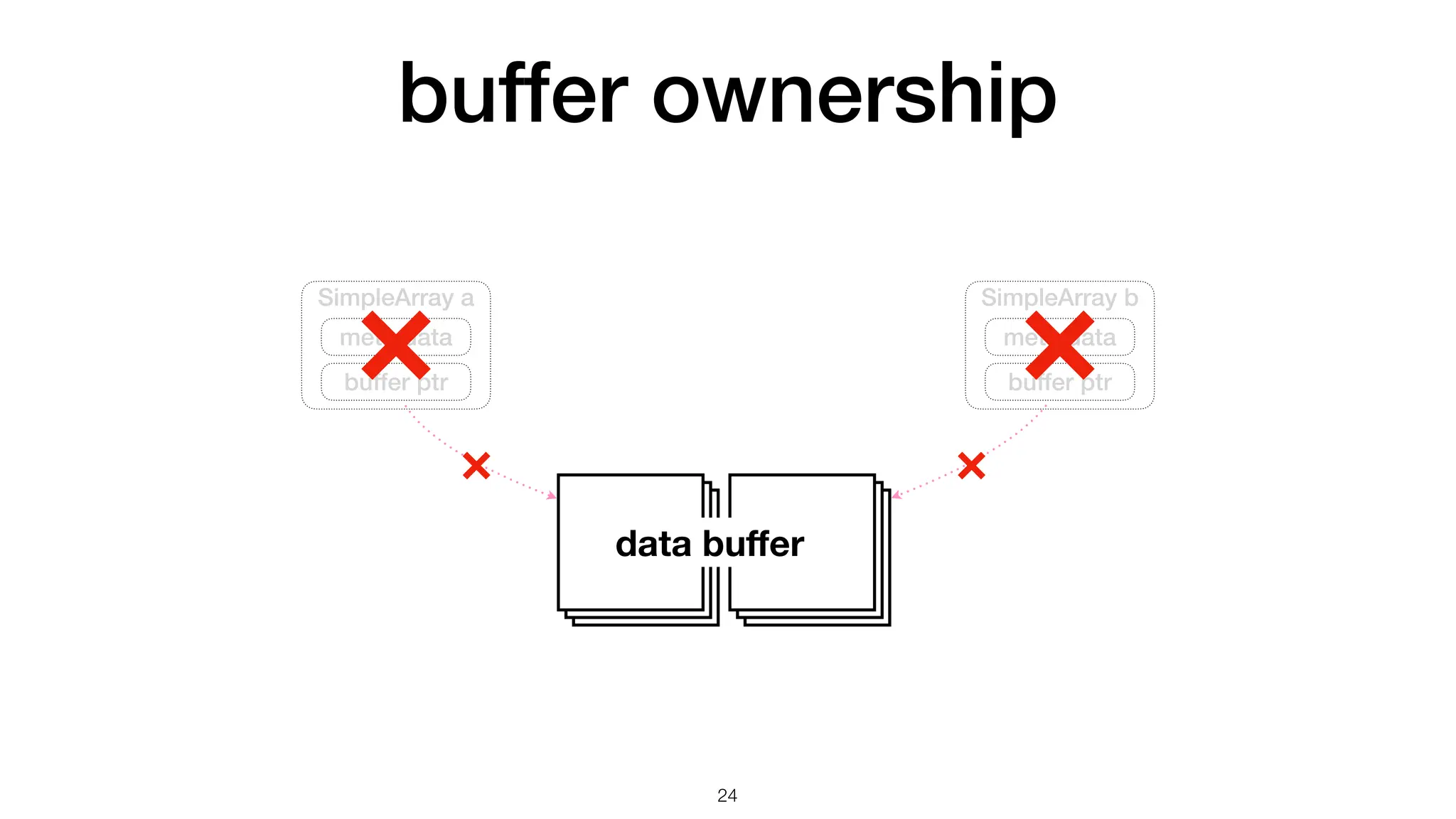
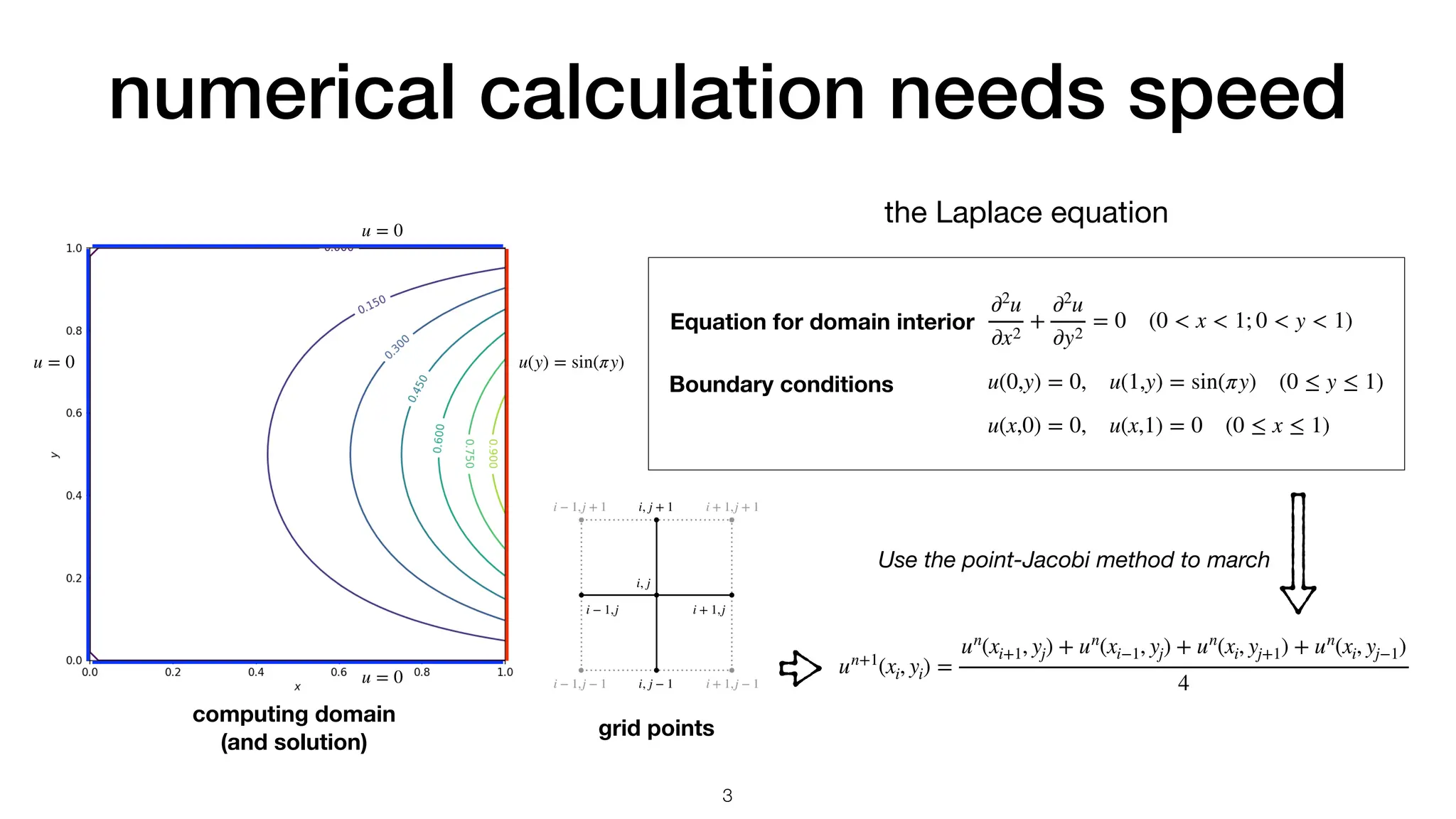
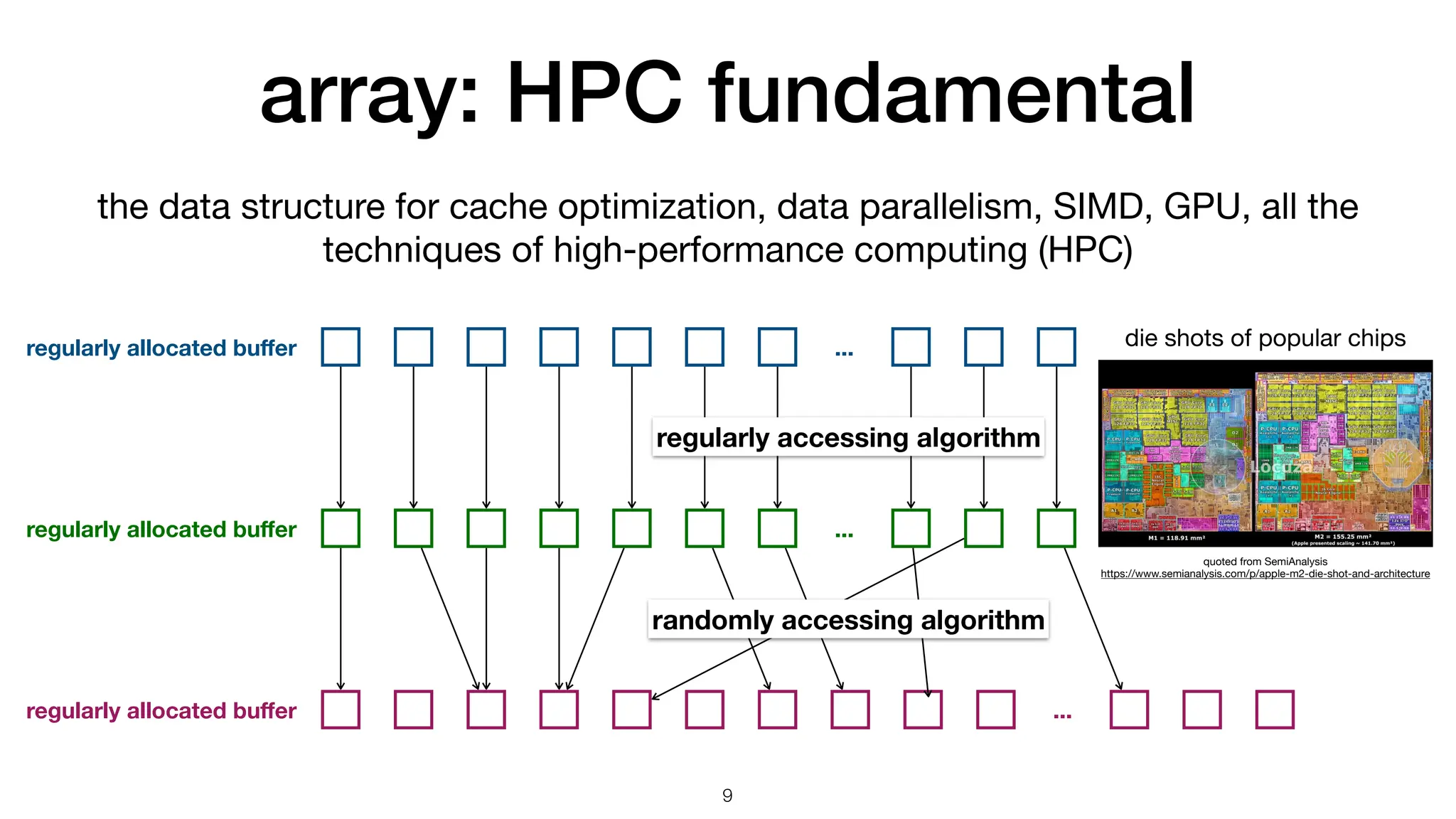
The document discusses the importance of speed in computational methods, particularly in relation to high-speed rail and numerical calculations. It provides an overview of the Point-Jacobi method for solving the Laplace equation, comparing the performance of Python and C++ implementations. It highlights the need for hybrid programming approaches that leverage Python for ease of use while utilizing C++ for performance-critical tasks in high-performance computing.


![Python is slow
for it in range(1, nx-1):
for jt in range(1, nx-1):
un[it,jt] = (u[it+1,jt] + u[it-1,jt] + u[it,jt+1] + u[it,jt-1]) / 4
un[1:nx-1,1:nx-1] = (u[2:nx,1:nx-1] + u[0:nx-2,1:nx-1] +
u[1:nx-1,2:nx] + u[1:nx-1,0:nx-2]) / 4
for (size_t it=1; it<nx-1; ++it)
{
for (size_t jt=1; jt<nx-1; ++jt)
{
un(it,jt) = (u(it+1,jt) + u(it-1,jt) + u(it,jt+1) + u(it,jt-1)) / 4;
}
}
Point-Jacobi method
Python nested loop
Numpy array
C++ nested loop
4.797s (1x)
0.055s (87x)
0.025s (192x)
un+1
(xi, yi) =
un
(xi+1, yj) + un
(xi−1, yj) + un
(xi, yj+1) + un
(xi, yj−1)
4
4](https://image.slidesharecdn.com/writepythonforspeedpyconapac20231027-231028212349-d3988144/75/Write-Python-for-Speed-4-2048.jpg)
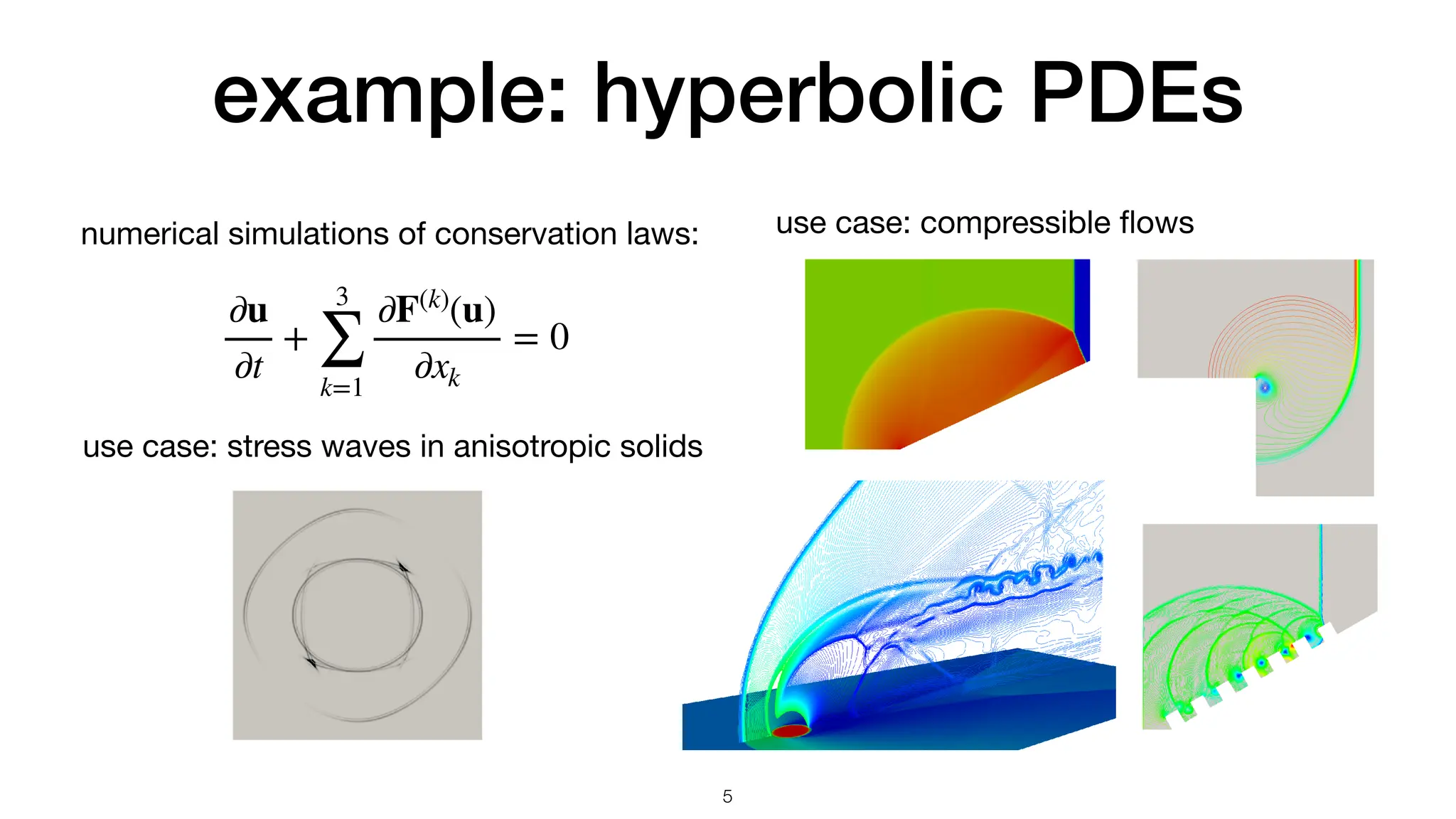
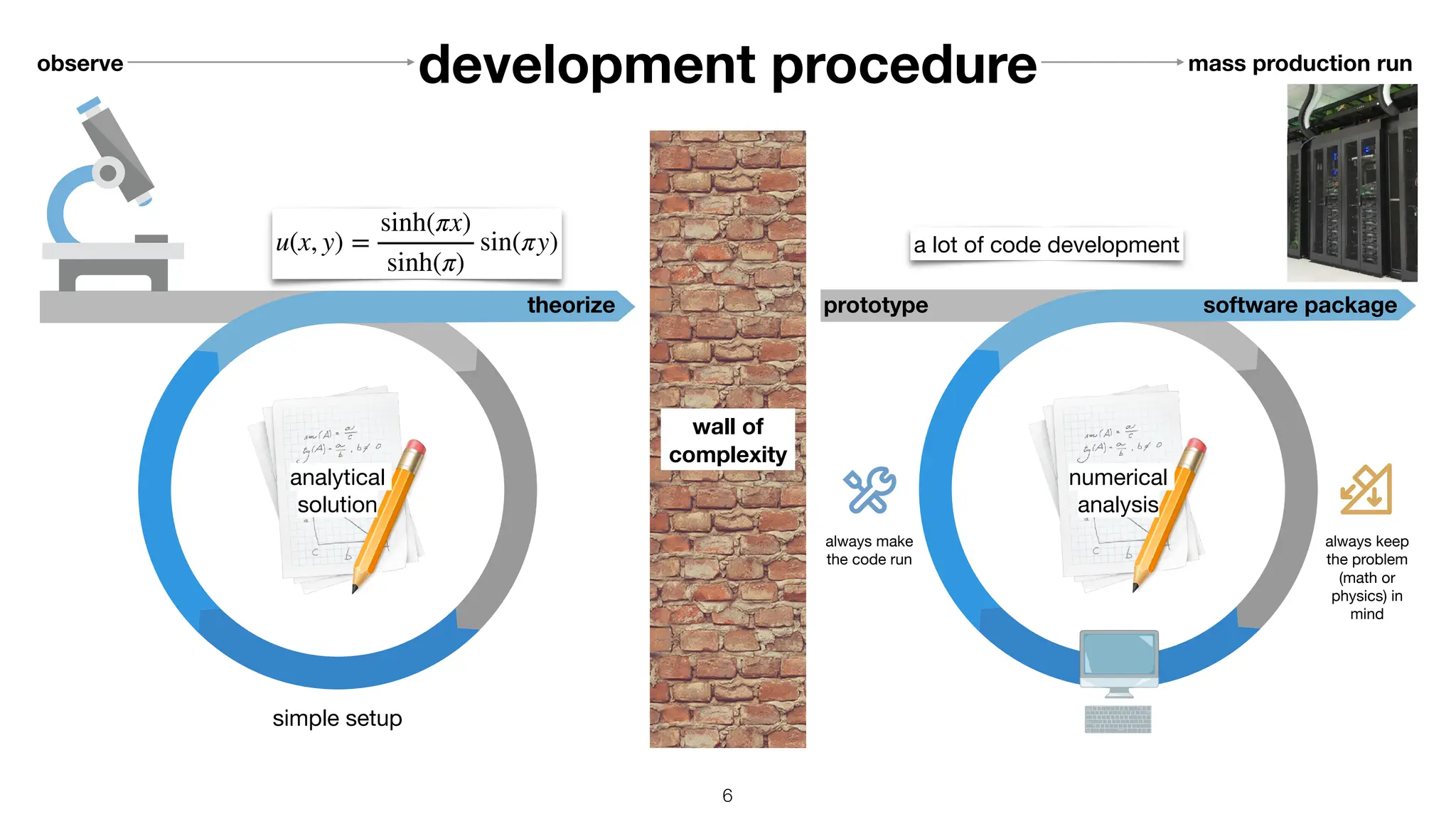
![machine determines speed
void calc_distance(
size_t const n
, double const * x
, double const * y
, double * r)
{
for (size_t i = 0 ; i < n ; ++i)
{
r[i] = std::sqrt(x[i]*x[i] + y[i]*y[i]);
}
}
vmovupd ymm0, ymmword [rsi + r9*8]
vmulpd ymm0, ymm0, ymm0
vmovupd ymm1, ymmword [rdx + r9*8]
vmulpd ymm1, ymm1, ymm1
vaddpd ymm0, ymm0, ymm1
vsqrtpd ymm0, ymm0
movupd xmm0, xmmword [rsi + r8*8]
mulpd xmm0, xmm0
movupd xmm1, xmmword [rdx + r8*8]
mulpd xmm1, xmm1
addpd xmm1, xmm0
sqrtpd xmm0, xmm1
AVX: 256-bit-wide vectorization SSE: 128-bit-wide vectorization
C++ code
8](https://image.slidesharecdn.com/writepythonforspeedpyconapac20231027-231028212349-d3988144/75/Write-Python-for-Speed-8-2048.jpg)
![quick prototype in Python:
something new and fragile
• thread pool for I/O? certainly
Python
• certainly not want to write
complicated C++ to prototype
• 64 threads on 32-core servers?
• consider to move it to C++
• data-parallel code on top of the
thread pool?
• time to go with TBB (thread-
building block) C++ library
from _thread import allocate_lock, start_new_thread
class ThreadPool(object):
"""Python prototype for I/O thread pool"""
def __init__(self, nthread):
# Worker callback
self.func = None
# Placeholders for managing data
self.__threadids = [None] * nthread
self.__threads = [None] * nthread
self.__returns = [None] * nthread
# Initialize thread managing data
for it in range(nthread):
mlck = allocate_lock(); mlck.acquire()
wlck = allocate_lock(); wlck.acquire()
tdata = [mlck, wlck, None, None]
self.__threads[it] = tdata
tid = start_new_thread(self.eventloop, (tdata,))
self.__threadids[it] = tid
11](https://image.slidesharecdn.com/writepythonforspeedpyconapac20231027-231028212349-d3988144/75/Write-Python-for-Speed-11-2048.jpg)
![quick prototype in Python:
something complex
i, j
i, j + 1
i, j − 1
i + 1,j
i − 1,j
i + 1,j + 1
i + 1,j − 1
i − 1,j + 1
i − 1,j − 1
di
ff
erence equation
def solve_python_loop():
u = uoriginal.copy() # Input from outer scope
un = u.copy() # Create the buffer for the next time step
converged = False
step = 0
# Outer loop.
while not converged:
step += 1
# Inner loops. One for x and the other for y.
for it in range(1, nx-1):
for jt in range(1, nx-1):
un[it,jt] = (u[it+1,jt] + u[it-1,jt] + u[it,jt+1] + u[it,jt-1]) / 4
norm = np.abs(un-u).max()
u[...] = un[...]
converged = True if norm < 1.e-5 else False
return u, step, norm
12
grid points
python nested loop implementing point-Jacobi method
u(xi, yj) =
u(xi+1, yj) + u(xi−1, yj) + u(xi, yj+1) + u(xi, yj−1)
4
point-Jacobi method
un+1
(xi, yi) =
un
(xi+1, yj) + un
(xi−1, yj) + un
(xi, yj+1) + un
(xi, yj−1)
4](https://image.slidesharecdn.com/writepythonforspeedpyconapac20231027-231028212349-d3988144/75/Write-Python-for-Speed-12-2048.jpg)
![production: many data many ways
13
fi
t scattered points to a polynomial
f(x) = a3x3
+ a2x2
+ a1x + a0
test many (like 1,000+) such data sets:
def plot_poly_fitted(i):
slct = (xdata>=i)&(xdata<(i+1))
sub_x = xdata[slct]
sub_y = ydata[slct]
poly = data_prep.fit_poly(sub_x, sub_y, 3)
print(poly)
poly = np.poly1d(poly)
xp = np.linspace(sub_x.min(), sub_x.max(), 100)
plt.plot(sub_x, sub_y, '.', xp, poly(xp), '-')
plot_poly_fitted(10)
(yeah, the data points seem to be too random to be represented by a polynomial)
// The rank of the linear map is (order+1).
modmesh::SimpleArray<double> matrix(std::vector<size_t>{order+1, order+1});
// Use the x coordinates to build the linear map for least-square
// regression.
for (size_t it=0; it<order+1; ++it)
{
for (size_t jt=0; jt<order+1; ++jt)
{
double & val = matrix(it, jt);
val = 0;
for (size_t kt=start; kt<stop; ++kt)
{
val += pow(xarr[kt], it+jt);
}
}
}
>>> with Timer():
>>> # Do the calculation for the 1000 groups of points.
>>> polygroup = np.empty((1000, 3), dtype='float64')
>>> for i in range(1000):
>>> # Use numpy to build the point group.
>>> slct = (xdata>=i)&(xdata<(i+1))
>>> sub_x = xdata[slct]
>>> sub_y = ydata[slct]
>>> polygroup[i,:] = data_prep.fit_poly(sub_x, sub_y, 2)
>>> with Timer():
>>> # Using numpy to build the point groups takes a lot of time.
>>> data_groups = []
>>> for i in range(1000):
>>> slct = (xdata>=i)&(xdata<(i+1))
>>> data_groups.append((xdata[slct], ydata[slct]))
>>> with Timer():
>>> # Fitting helper runtime is much less than building the point groups.
>>> polygroup = np.empty((1000, 3), dtype='float64')
>>> for it, (sub_x, sub_y) in enumerate(data_groups):
>>> polygroup[it,:] = data_prep.fit_poly(sub_x, sub_y, 2)
Wall time: 1.49671 s
Wall time: 1.24653 s
Wall time: 0.215859 s
prepare data
fi
t polynomials](https://image.slidesharecdn.com/writepythonforspeedpyconapac20231027-231028212349-d3988144/75/Write-Python-for-Speed-13-2048.jpg)
![problem of speedup
14
>>> with Timer():
>>> # Using numpy to build the point groups takes a lot of time.
>>> data_groups = []
>>> for i in range(1000):
>>> slct = (xdata>=i)&(xdata<(i+1))
>>> data_groups.append((xdata[slct], ydata[slct]))
Wall time: 1.24653 s
>>> with Timer():
>>> # Fitting helper runtime is much less than building the point groups.
>>> polygroup = np.empty((1000, 3), dtype='float64')
>>> for it, (sub_x, sub_y) in enumerate(data_groups):
>>> polygroup[it,:] = data_prep.fit_poly(sub_x, sub_y, 2)
Wall time: 0.215859 s
>>> with Timer():
>>> rbatch = data_prep.fit_polys(xdata, ydata, 2)
Wall time: 0.21058 s
/**
* This function calculates the least-square regression of multiple sets of
* point clouds to the corresponding polynomial functions of a given order.
*/
modmesh::SimpleArray<double> fit_polys
(
modmesh::SimpleArray<double> const & xarr
, modmesh::SimpleArray<double> const & yarr
, size_t order
)
{
size_t xmin = std::floor(*std::min_element(xarr.begin(), xarr.end()));
size_t xmax = std::ceil(*std::max_element(xarr.begin(), xarr.end()));
size_t ninterval = xmax - xmin;
modmesh::SimpleArray<double> lhs(std::vector<size_t>{ninterval, order+1});
std::fill(lhs.begin(), lhs.end(), 0); // sentinel.
size_t start=0;
for (size_t it=0; it<xmax; ++it)
{
// NOTE: We take advantage of the intrinsic features of the input data
// to determine the grouping. This is ad hoc and hard to maintain. We
// play this trick to demonstrate a hackish way of performing numerical
// calculation.
size_t stop;
for (stop=start; stop<xarr.size(); ++stop)
{
if (xarr[stop]>=it+1) { break; }
}
// Use the single polynomial helper function.
auto sub_lhs = fit_poly(xarr, yarr, start, stop, order);
for (size_t jt=0; jt<order+1; ++jt)
{
lhs(it, jt) = sub_lhs[jt];
}
start = stop;
}
return lhs;
}
prepare data and loop in Python:
prepare data and loop in C++:
C++ wins so much, but we lose
fl
exibility!
// NOTE: We take advantage of the intrinsic features of the input data
// to determine the grouping. This is ad hoc and hard to maintain. We
// play this trick to demonstrate a hackish way of performing numerical
// calculation.](https://image.slidesharecdn.com/writepythonforspeedpyconapac20231027-231028212349-d3988144/75/Write-Python-for-Speed-14-2048.jpg)

 -> decltype(auto)
{ return self.NAME(); })
(*this)
// geometry arrays
MM_DECL_ARRAY(ndcrd)
MM_DECL_ARRAY(fccnd)
MM_DECL_ARRAY(fcnml)
MM_DECL_ARRAY(fcara)
MM_DECL_ARRAY(clcnd)
MM_DECL_ARRAY(clvol)
// meta arrays
MM_DECL_ARRAY(fctpn)
MM_DECL_ARRAY(cltpn)
MM_DECL_ARRAY(clgrp)
// connectivity arrays
MM_DECL_ARRAY(fcnds)
MM_DECL_ARRAY(fccls)
MM_DECL_ARRAY(clnds)
MM_DECL_ARRAY(clfcs)
MM_DECL_ARRAY(ednds);
#undef MM_DECL_ARRAY
# Construct the data object
mh = modmesh.StaticMesh(
ndim=3, nnode=4, nface=4, ncell=1)
# Set the data
mh.ndcrd.ndarray[:, :] =
(0, 0, 0), (0, 1, 0), (-1, 1, 0), (0, 1, 1)
mh.cltpn.ndarray[:] = modmesh.StaticMesh.TETRAHEDRON
mh.clnds.ndarray[:, :5] = [(4, 0, 1, 2, 3)]
# Calculate internal by the input data
# to build up the object
mh.build_interior()
np.testing.assert_almost_equal(
mh.fccnd,
[[-0.3333333, 0.6666667, 0. ],
[ 0. , 0.6666667, 0.3333333],
[-0.3333333, 0.6666667, 0.3333333],
[-0.3333333, 1. , 0.3333333]])
mesh shape information
data arrays
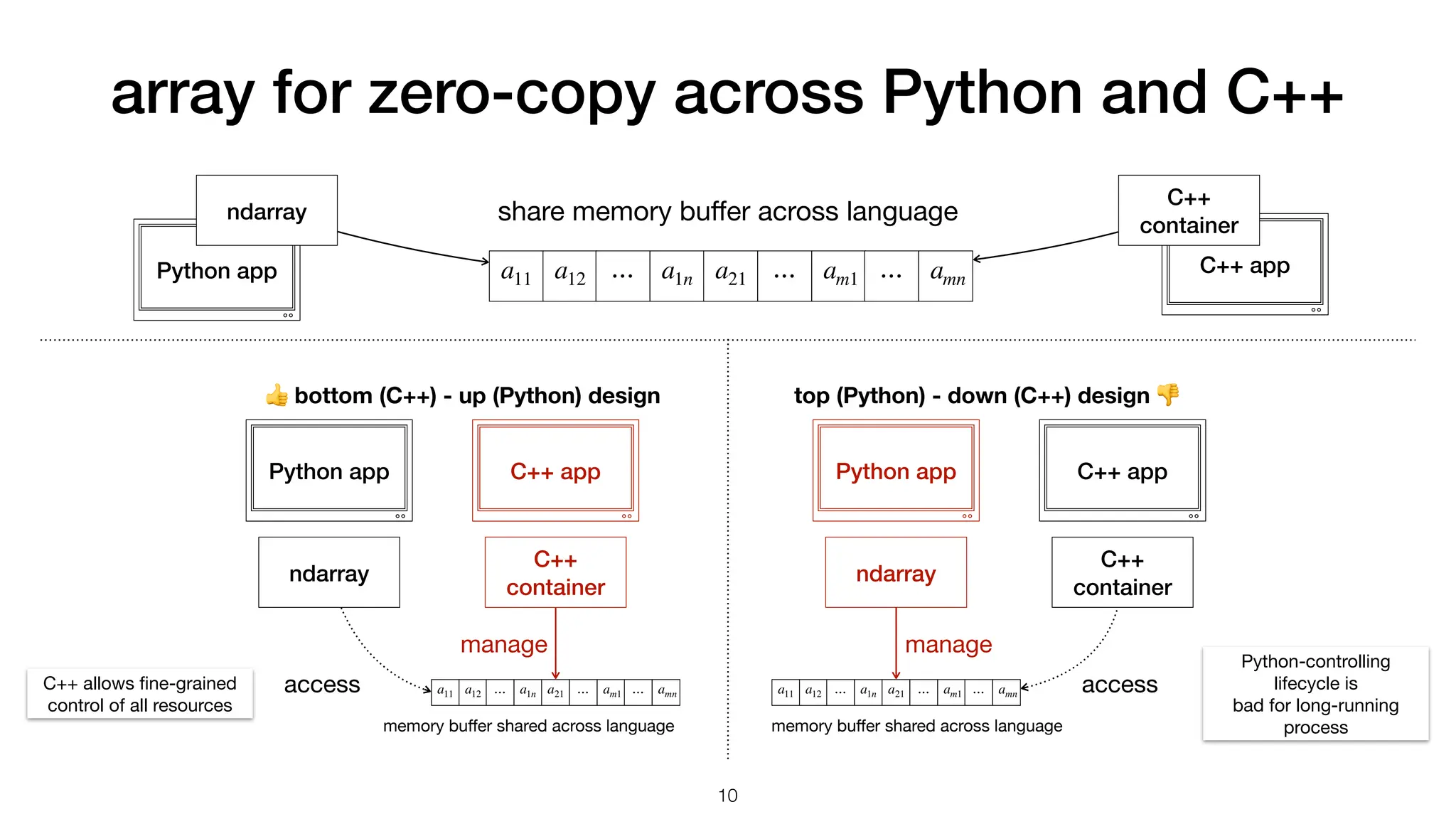
data arrays will be very large: gigabytes in memory
use data in python
C++ library pybind11 wrapper
16](https://image.slidesharecdn.com/writepythonforspeedpyconapac20231027-231028212349-d3988144/75/Write-Python-for-Speed-16-2048.jpg)
![test in Python
def test_2d_trivial_triangles(self):
mh = modmesh.StaticMesh(ndim=2, nnode=4, nface=0, ncell=3)
mh.ndcrd.ndarray[:, :] = (0, 0), (-1, -1), (1, -1), (0, 1)
mh.cltpn.ndarray[:] = modmesh.StaticMesh.TRIANGLE
mh.clnds.ndarray[:, :4] = (3, 0, 1, 2), (3, 0, 2, 3), (3, 0, 3, 1)
self._check_shape(mh, ndim=2, nnode=4, nface=0, ncell=3,
nbound=0, ngstnode=0, ngstface=0, ngstcell=0,
nedge=0)
# Test build interior data.
mh.build_interior(_do_metric=False, _build_edge=False)
self._check_shape(mh, ndim=2, nnode=4, nface=6, ncell=3,
nbound=0, ngstnode=0, ngstface=0, ngstcell=0,
nedge=0)
mh.build_interior() # _do_metric=True, _build_edge=True
self._check_shape(mh, ndim=2, nnode=4, nface=6, ncell=3,
nbound=0, ngstnode=0, ngstface=0, ngstcell=0,
nedge=6)
np.testing.assert_almost_equal(
mh.fccnd,
[[-0.5, -0.5], [0.0, -1.0], [0.5, -0.5],
[0.5, 0.0], [0.0, 0.5], [-0.5, 0.0]])
np.testing.assert_almost_equal(
mh.fcnml,
[[-0.7071068, 0.7071068], [0.0, -1.0], [0.7071068, 0.7071068],
[0.8944272, 0.4472136], [-1.0, -0.0], [-0.8944272, 0.4472136]])
np.testing.assert_almost_equal(
mh.fcara, [1.4142136, 2.0, 1.4142136, 2.236068, 1.0, 2.236068])
np.testing.assert_almost_equal(
mh.clcnd, [[0.0, -0.6666667], [0.3333333, 0.0], [-0.3333333, 0.0]])
np.testing.assert_almost_equal(
mh.clvol, [1.0, 0.5, 0.5])
17
namespace py = pybind11;
(*this)
// shape data
.def_property_readonly("ndim", &wrapped_type::ndim)
.def_property_readonly("nnode", &wrapped_type::nnode)
.def_property_readonly("nface", &wrapped_type::nface)
.def_property_readonly("ncell", &wrapped_type::ncell)
.def_property_readonly("nbound", &wrapped_type::nbound)
.def_property_readonly("ngstnode", &wrapped_type::ngstnode)
.def_property_readonly("ngstface", &wrapped_type::ngstface)
.def_property_readonly("ngstcell", &wrapped_type::ngstcell)
#define MM_DECL_ARRAY(NAME) .expose_SimpleArray(
#NAME,
[](wrapped_type & self) -> decltype(auto){ return self.NAME(); })
(*this)
// geometry arrays
MM_DECL_ARRAY(ndcrd)
MM_DECL_ARRAY(fccnd)
MM_DECL_ARRAY(fcnml)
MM_DECL_ARRAY(fcara)
MM_DECL_ARRAY(clcnd)
MM_DECL_ARRAY(clvol)
// meta arrays
MM_DECL_ARRAY(fctpn)
MM_DECL_ARRAY(cltpn)
MM_DECL_ARRAY(clgrp)
// connectivity arrays
MM_DECL_ARRAY(fcnds)
MM_DECL_ARRAY(fccls)
MM_DECL_ARRAY(clnds)
MM_DECL_ARRAY(clfcs)
MM_DECL_ARRAY(ednds);
#undef MM_DECL_ARRAY
wrapping to Python enables fast testing development may be done as writing tests!
# Construct the data object
mh = modmesh.StaticMesh(
ndim=3, nnode=4, nface=4, ncell=1)
# Set the data
mh.ndcrd.ndarray[:, :] =
(0, 0, 0), (0, 1, 0), (-1, 1, 0), (0, 1, 1)
mh.cltpn.ndarray[:] = modmesh.StaticMesh.TETRAHEDRON
mh.clnds.ndarray[:, :5] = [(4, 0, 1, 2, 3)]
# Calculate internal by the input data
# to build up the object
mh.build_interior()
np.testing.assert_almost_equal(
mh.fccnd,
[[-0.3333333, 0.6666667, 0. ],
[ 0. , 0.6666667, 0.3333333],
[-0.3333333, 0.6666667, 0.3333333],
[-0.3333333, 1. , 0.3333333]])](https://image.slidesharecdn.com/writepythonforspeedpyconapac20231027-231028212349-d3988144/75/Write-Python-for-Speed-17-2048.jpg)
![develop SimpleArray in C++
SimpleArray std::vector
SimpleArray is
fi
xed size 👍
• only allocate memory on
construction
std::vector is variable size 👎
• bu
ff
er may be invalidated
• implicit memory allocation
(reallocation)
multi-dimensional access 👍
operator()
one-dimensional access 👎
operator[]
19
C++
container
ndarray
manage
access a11 a12 ⋯ a1n a21 ⋯ am1 ⋯ amn
memory bu
ff
er
make it yourself get it free from STL](https://image.slidesharecdn.com/writepythonforspeedpyconapac20231027-231028212349-d3988144/75/Write-Python-for-Speed-19-2048.jpg)