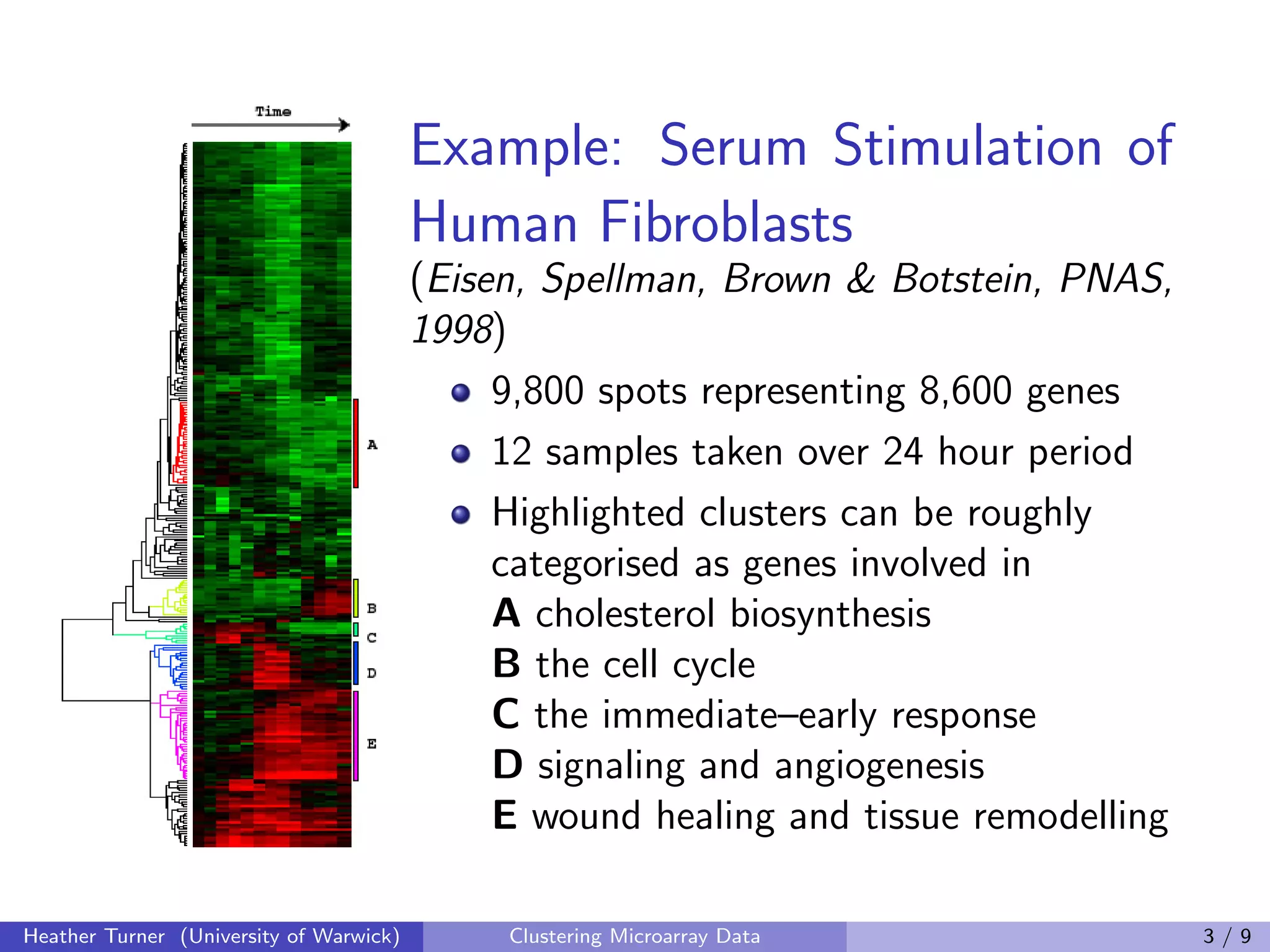
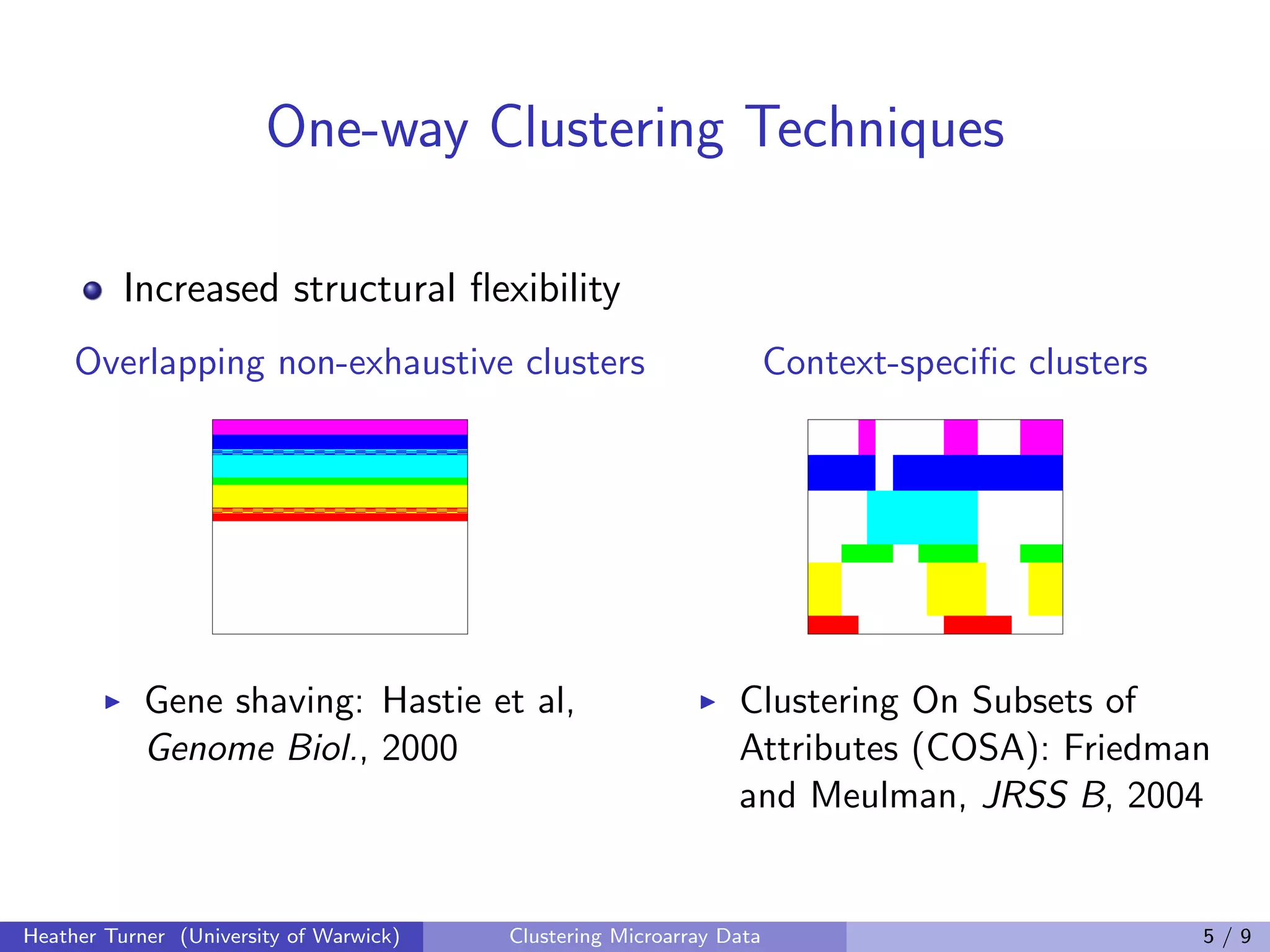
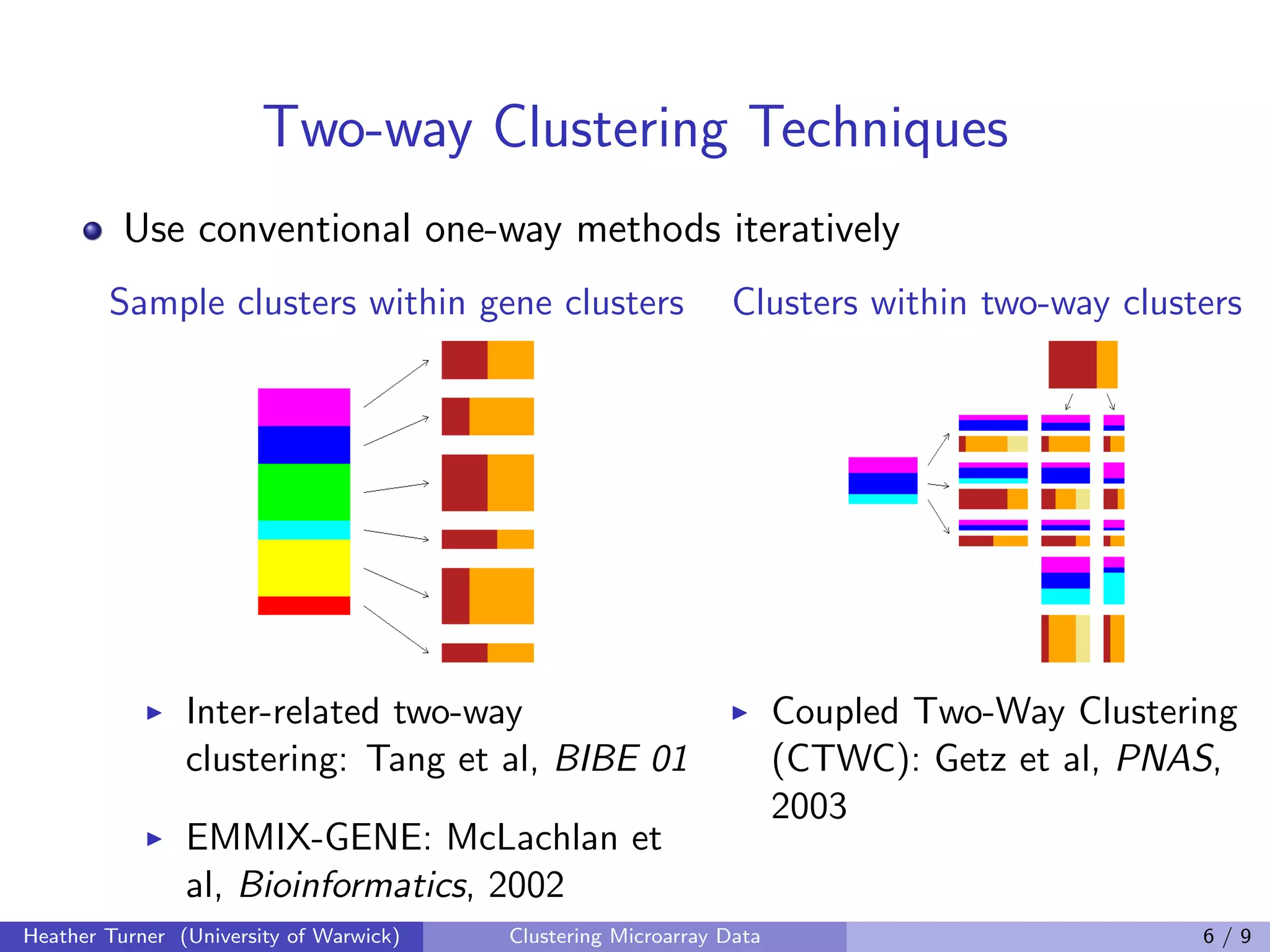
This document discusses various techniques for clustering microarray data, including one-way, two-way, co-clustering, and biclustering techniques. It provides examples of methods such as gene shaving, COSA, coupled two-way clustering, spectral bi-clustering, and SAMBA. While many novel clustering methods have been developed, the document notes that few are widely used in practice and there has been little work evaluating the performance of different techniques. Development of clustering methods for microarray data continues.