Aspergillus fumigatus strains that evolve resistance to the agrochemical fungicide ipflufenoquin in vitro are also resistant to olorofim
- PMID: 38151646
- PMCID: PMC10769868
- DOI: 10.1038/s41564-023-01542-4
Aspergillus fumigatus strains that evolve resistance to the agrochemical fungicide ipflufenoquin in vitro are also resistant to olorofim
Abstract
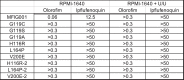
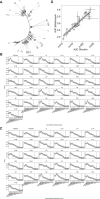
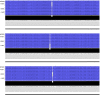
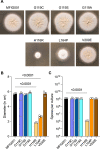
Widespread use of azole antifungals in agriculture has been linked to resistance in the pathogenic fungus Aspergillus fumigatus. We show that exposure of A. fumigatus to the agrochemical fungicide, ipflufenoquin, in vitro can select for strains that are resistant to olorofim, a first-in-class clinical antifungal with the same mechanism of action. Resistance is caused by non-synonymous mutations within the target of ipflufenoquin/olorofim activity, dihydroorotate dehydrogenase (DHODH), and these variants have no overt growth defects.
© 2023. The Author(s).
Conflict of interest statement
M. J. Bromley is a former employee of F2G Ltd (until 2007), the company that developed the antifungal olorofim. He has received historic funding for PhD studentships from F2G Ltd but has no current financial interest in F2G Ltd. M.B. and J.D.O. are employed by F2G Ltd. Experiments carried out at the University of Manchester were done independently and without input or incumbrance from F2G Ltd. F2G Ltd contribution was solely in relation to the IC50 analysis of AfDHODH variants. All remaining authors declare no competing interests.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

