Genome-wide analysis of heat stress-stimulated transposon mobility in the human fungal pathogen Cryptococcus deneoformans
- PMID: 36669112
- PMCID: PMC9942834
- DOI: 10.1073/pnas.2209831120
Genome-wide analysis of heat stress-stimulated transposon mobility in the human fungal pathogen Cryptococcus deneoformans
Abstract
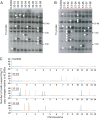
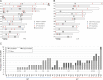
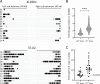
We recently reported transposon mutagenesis as a significant driver of spontaneous mutations in the human fungal pathogen Cryptococcus deneoformans during murine infection. Mutations caused by transposable element (TE) insertion into reporter genes were dramatically elevated at high temperatures (37° vs. 30°) in vitro, suggesting that heat stress stimulates TE mobility in the Cryptococcus genome. To explore the genome-wide impact of TE mobilization, we generated transposon accumulation lines by in vitro passage of C. deneoformans strain XL280α for multiple generations at both 30° and at the host-relevant temperature of 37°. Utilizing whole-genome sequencing, we identified native TE copies and mapped multiple de novo TE insertions in these lines. Movements of the T1 DNA transposon occurred at both temperatures with a strong bias for insertion between gene-coding regions. By contrast, the Tcn12 retrotransposon integrated primarily within genes and movement occurred exclusively at 37°. In addition, we observed a dramatic amplification in copy number of the Cnl1 (Cryptococcus neoformans LINE-1) retrotransposon in subtelomeric regions under heat-stress conditions. Comparing TE mutations to other sequence variations detected in passaged lines, the increase in genomic changes at elevated temperatures was primarily due to mobilization of the retroelements Tcn12 and Cnl1. Finally, we found multiple TE movements (T1, Tcn12, and Cnl1) in the genomes of single C. deneoformans isolates recovered from infected mice, providing evidence that mobile elements are likely to facilitate microevolution and rapid adaptation during infection.
Keywords: Cryptococcus; fungal pathogen; heat stress; temperature; transposons.
Conflict of interest statement
The authors have additional information to disclose, Author J.H. and reviewer E.H.S. are co-authors on a 2020 review together (DOI:
Figures


References
-
- Hagen F., et al. , Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal. Genet. Biol. 78, 16–48 (2015). - PubMed
-
- Lin X., Heitman J., The biology of the Cryptococcus neoformans species complex. Annu. Rev. Microbiol. 60, 69–105 (2006). - PubMed
-
- Casadevall A., Kwon-Chung K. J., Perfect J. R., Kozel T. R., Heitman J., Cryptococcus: From Human Pathogen to Model Yeast (ASM Press, 2011).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

