Transposon mobilization in the human fungal pathogen Cryptococcus is mutagenic during infection and promotes drug resistance in vitro
- PMID: 32303657
- PMCID: PMC7211991
- DOI: 10.1073/pnas.2001451117
Transposon mobilization in the human fungal pathogen Cryptococcus is mutagenic during infection and promotes drug resistance in vitro
Abstract
When transitioning from the environment, pathogenic microorganisms must adapt rapidly to survive in hostile host conditions. This is especially true for environmental fungi that cause opportunistic infections in immunocompromised patients since these microbes are not well adapted human pathogens. Cryptococcus species are yeastlike fungi that cause lethal infections, especially in HIV-infected patients. Using Cryptococcus deneoformans in a murine model of infection, we examined contributors to drug resistance and demonstrated that transposon mutagenesis drives the development of 5-fluoroorotic acid (5FOA) resistance. Inactivation of target genes URA3 or URA5 primarily reflected the insertion of two transposable elements (TEs): the T1 DNA transposon and the TCN12 retrotransposon. Consistent with in vivo results, increased rates of mutagenesis and resistance to 5FOA and the antifungal drugs rapamycin/FK506 (rap/FK506) and 5-fluorocytosine (5FC) were found when Cryptococcus was incubated at 37° compared to 30° in vitro, a condition that mimics the temperature shift that occurs during the environment-to-host transition. Inactivation of the RNA interference (RNAi) pathway, which suppresses TE movement in many organisms, was not sufficient to elevate TE movement at 30° to the level observed at 37°. We propose that temperature-dependent TE mobilization in Cryptococcus is an important mechanism that enhances microbial adaptation and promotes pathogenesis and drug resistance in the human host.
Keywords: Cryptococcus; drug resistance; fungal pathogen; temperature; transposons.
Conflict of interest statement
Competing interest statement: A.C. and J.H. are coauthors on a forthcoming position paper. They have not collaborated directly.
Figures
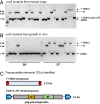


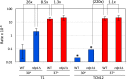
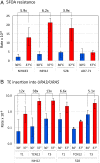
References
-
- Hagen F., et al. , Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet. Biol. 78, 16–48 (2015). - PubMed
-
- Lin X., Heitman J., The biology of the Cryptococcus neoformans species complex. Annu. Rev. Microbiol. 60, 69–105 (2006). - PubMed
-
- Dromer F., Mathoulin S., Dupont B., Letenneur L., Ronin O., Individual and environmental factors associated with infection due to Cryptococcus neoformans serotype D. French Cryptococcosis Study Group. Clin. Infect. Dis. 23, 91–96 (1996). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

