FPR1 is the plague receptor on host immune cells
- PMID: 31534221
- PMCID: PMC6776691
- DOI: 10.1038/s41586-019-1570-z
FPR1 is the plague receptor on host immune cells
Abstract
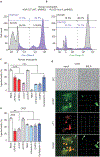
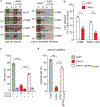
The causative agent of plague, Yersinia pestis, uses a type III secretion system to selectively destroy immune cells in humans, thus enabling Y. pestis to reproduce in the bloodstream and be transmitted to new hosts through fleabites. The host factors that are responsible for the selective destruction of immune cells by plague bacteria are unknown. Here we show that LcrV, the needle cap protein of the Y. pestis type III secretion system, binds to the N-formylpeptide receptor (FPR1) on human immune cells to promote the translocation of bacterial effectors. Plague infection in mice is characterized by high mortality; however, Fpr1-deficient mice have increased survival and antibody responses that are protective against plague. We identified FPR1R190W as a candidate resistance allele in humans that protects neutrophils from destruction by the Y. pestis type III secretion system. Thus, FPR1 is a plague receptor on immune cells in both humans and mice, and its absence or mutation provides protection against Y. pestis. Furthermore, plague selection of FPR1 alleles appears to have shaped human immune responses towards other infectious diseases and malignant neoplasms.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
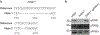


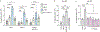
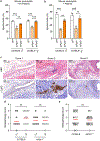

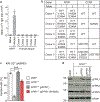
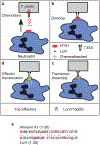
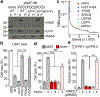
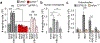
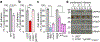

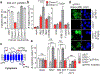
References
-
- Andrades Valtueña A et al. The stone age plague and its persistence in Eurasia. Curr Biol. 27, 3683–3691 (2017). - PubMed
-
- Benedictow OJ The Black Death 1346–1353: The Complete History. (Boydell Press, 2004).
-
- Samson M et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382, 722–725 (1996). - PubMed
-
- Liu R et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86, 367–377 (1996). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

