Identification and characterization of Sr13, a tetraploid wheat gene that confers resistance to the Ug99 stem rust race group
- PMID: 29078294
- PMCID: PMC5692537
- DOI: 10.1073/pnas.1706277114
Identification and characterization of Sr13, a tetraploid wheat gene that confers resistance to the Ug99 stem rust race group
Abstract
The Puccinia graminis f. sp. tritici (Pgt) Ug99 race group is virulent to most stem rust resistance genes currently deployed in wheat and poses a threat to global wheat production. The durum wheat (Triticum turgidum ssp. durum) gene Sr13 confers resistance to Ug99 and other virulent races, and is more effective at high temperatures. Using map-based cloning, we delimited a candidate region including two linked genes encoding coiled-coil nucleotide-binding leucine-rich repeat proteins designated CNL3 and CNL13. Three independent truncation mutations identified in each of these genes demonstrated that only CNL13 was required for Ug99 resistance. Transformation of an 8-kb genomic sequence including CNL13 into the susceptible wheat variety Fielder was sufficient to confer resistance to Ug99, confirming that CNL13 is Sr13CNL13 transcripts were slightly down-regulated 2-6 days after Pgt inoculation and were not affected by temperature. By contrast, six pathogenesis-related (PR) genes were up-regulated at high temperatures only when both Sr13 and Pgt were present, suggesting that they may contribute to the high temperature resistance mechanism. We identified three Sr13-resistant haplotypes, which were present in one-third of cultivated emmer and durum wheats but absent in most tested common wheats (Triticum aestivum). These results suggest that Sr13 can be used to improve Ug99 resistance in a large proportion of modern wheat cultivars. To accelerate its deployment, we developed a diagnostic marker for Sr13 The identification of Sr13 expands the number of Pgt-resistance genes that can be incorporated into multigene transgenic cassettes to control this devastating disease.
Keywords: CC-NBS-LRR; Ug99; durum wheat; resistance genes; stem rust.
Copyright © 2017 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
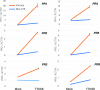
References
-
- Pretorius ZA, Singh RP, Wagoire WW, Payne TS. Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis f. sp. tritici in Uganda. Plant Dis. 2000;84:203. - PubMed
-
- Jin Y, et al. Detection of virulence to resistance gene Sr24 within race TTKS of Puccinia graminis f. sp tritici. Plant Dis. 2008;92:923–926. - PubMed
-
- Jin Y, et al. Detection of virulence to resistance gene Sr36 within the TTKS race lineage of Puccinia graminis f. sp tritici. Plant Dis. 2009;93:367–370. - PubMed
-
- Pretorius ZA, Szabo G, Boshoff WHP, Herselman L, Visser B. First report of a new TTKSF race of wheat stem rust (Puccinia graminis f. sp. tritici) in South Africa and Zimbabwe. A. Pretorius. Plant Dis. 2012;96:590. - PubMed
-
- Rouse MN, et al. Characterization of Sr9h, a wheat stem rust resistance allele effective to Ug99. Theor Appl Genet. 2014;127:1681–1688. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

