Helminth infection promotes colonization resistance via type 2 immunity
- PMID: 27080105
- PMCID: PMC4905769
- DOI: 10.1126/science.aaf3229
Helminth infection promotes colonization resistance via type 2 immunity
Abstract
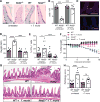
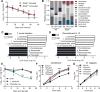
Increasing incidence of inflammatory bowel diseases, such as Crohn's disease, in developed nations is associated with changes to the microbial environment, such as decreased prevalence of helminth colonization and alterations to the gut microbiota. We find that helminth infection protects mice deficient in the Crohn's disease susceptibility gene Nod2 from intestinal abnormalities by inhibiting colonization by an inflammatory Bacteroides species. Resistance to Bacteroides colonization was dependent on type 2 immunity, which promoted the establishment of a protective microbiota enriched in Clostridiales. Additionally, we show that individuals from helminth-endemic regions harbor a similar protective microbiota and that deworming treatment reduced levels of Clostridiales and increased Bacteroidales. These results support a model of the hygiene hypothesis in which certain individuals are genetically susceptible to the consequences of a changing microbial environment.
Copyright © 2016, American Association for the Advancement of Science.
Figures


Comment in
-
IBD: Parasites promote protective microbiota.Nat Rev Gastroenterol Hepatol. 2016 Jun;13(6):316. doi: 10.1038/nrgastro.2016.73. Epub 2016 May 5. Nat Rev Gastroenterol Hepatol. 2016. PMID: 27147492 No abstract available.
-
Helminths and Intestinal Flora Team Up to Improve Gut Health.Trends Parasitol. 2016 Sep;32(9):664-666. doi: 10.1016/j.pt.2016.05.006. Epub 2016 May 24. Trends Parasitol. 2016. PMID: 27234811
-
Intimate gut interactions: helminths and the microbiota.Cell Res. 2016 Aug;26(8):861-2. doi: 10.1038/cr.2016.72. Epub 2016 Jun 14. Cell Res. 2016. PMID: 27297236 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI093811/AI/NIAID NIH HHS/United States
- UL1 TR001445/TR/NCATS NIH HHS/United States
- DK093668/DK/NIDDK NIH HHS/United States
- P30 CA016087/CA/NCI NIH HHS/United States
- AI007180/AI/NIAID NIH HHS/United States
- R01 AI107588/AI/NIAID NIH HHS/United States
- R01 DK093668/DK/NIDDK NIH HHS/United States
- AI107588/AI/NIAID NIH HHS/United States
- UL1 TR00038/TR/NCATS NIH HHS/United States
- DK090989/DK/NIDDK NIH HHS/United States
- T32 AI007180/AI/NIAID NIH HHS/United States
- R01 DK103788/DK/NIDDK NIH HHS/United States
- AI093811/AI/NIAID NIH HHS/United States
- DK103788/DK/NIDDK NIH HHS/United States
- UL1 TR000038/TR/NCATS NIH HHS/United States
- P30CA016087/CA/NCI NIH HHS/United States
- HL123340/HL/NHLBI NIH HHS/United States
- R01 DK090989/DK/NIDDK NIH HHS/United States
- R01 HL123340/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

