Genomic Insights into a New Citrobacter koseri Strain Revealed Gene Exchanges with the Virulence-Associated Yersinia pestis pPCP1 Plasmid
- PMID: 27014253
- PMCID: PMC4793686
- DOI: 10.3389/fmicb.2016.00340
Genomic Insights into a New Citrobacter koseri Strain Revealed Gene Exchanges with the Virulence-Associated Yersinia pestis pPCP1 Plasmid
Abstract
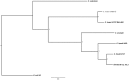
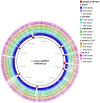
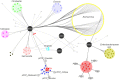
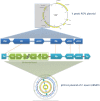
The history of infectious diseases raised the plague as one of the most devastating for human beings. Far too often considered an ancient disease, the frequent resurgence of the plague has led to consider it as a reemerging disease in Madagascar, Algeria, Libya, and Congo. The genetic factors associated with the pathogenicity of Yersinia pestis, the causative agent of the plague, involve the acquisition of the pPCP1 plasmid that promotes host invasion through the expression of the virulence factor Pla. The surveillance of plague foci after the 2003 outbreak in Algeria resulted in a positive detection of the specific pla gene of Y. pestis in rodents. However, the phenotypic characterization of the isolate identified a Citrobacter koseri. The comparative genomics of our sequenced C. koseri URMITE genome revealed a mosaic gene structure resulting from the lifestyle of our isolate and provided evidence for gene exchanges with different enteric bacteria. The most striking was the acquisition of a continuous 2 kb genomic fragment containing the virulence factor Pla of the Y. pestis pPCP1 plasmid; however, the subcutaneous injection of the CKU strain in mice did not produce any pathogenic effect. Our findings demonstrate that fast molecular detection of plague using solely the pla gene is unsuitable and should rather require Y. pestis gene marker combinations. We also suggest that the evolutionary force that might govern the expression of pathogenicity can occur through the acquisition of virulence genes but could also require the loss or the inactivation of resident genes such as antivirulence genes.
Keywords: Citrobacter koseri; bioinformatics; genomics and evolution; plague pathogenesis; virulence factors.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

