Ecological robustness of the gut microbiota in response to ingestion of transient food-borne microbes
- PMID: 26953599
- PMCID: PMC4989305
- DOI: 10.1038/ismej.2016.13
Ecological robustness of the gut microbiota in response to ingestion of transient food-borne microbes
Abstract
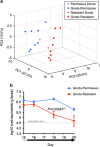
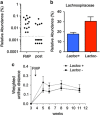
Resident gut microbes co-exist with transient bacteria to form the gut microbiota. Despite increasing evidence suggesting a role for transient microbes on gut microbiota function, the interplay between resident and transient members of this microbial community is poorly defined. We aimed to determine the extent to which a host's autochthonous gut microbiota influences niche permissivity to transient bacteria using a fermented milk product (FMP) as a vehicle for five food-borne bacterial strains. Using conventional and gnotobiotic rats and gut microbiome analyses (16S rRNA genes pyrosequencing and reverse transcription qPCR), we demonstrated that the clearance kinetics of one FMP bacterium, Lactococcus lactis CNCM I-1631, were dependent on the structure of the resident gut microbiota. Susceptibility of the resident gut microbiota to modulation by FMP intervention correlated with increased persistence of L. lactis. We also observed gut microbiome configurations that were associated with altered stability upon exposure to transient bacteria. Our study supports the concept that allochthonous bacteria have transient and subject-specific effects on the gut microbiome that can be leveraged to re-engineer the gut microbiome and improve dysbiosis-related diseases.
Figures


References
-
- Anderson MJ, Willis TJ. (2003). Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology 84: 511–525.
-
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H et al. (2013). Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500: 232–236. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

