Eighteenth century Yersinia pestis genomes reveal the long-term persistence of an historical plague focus
- PMID: 26795402
- PMCID: PMC4798955
- DOI: 10.7554/eLife.12994
Eighteenth century Yersinia pestis genomes reveal the long-term persistence of an historical plague focus
Abstract
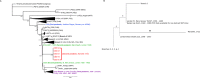
The 14th-18th century pandemic of Yersinia pestis caused devastating disease outbreaks in Europe for almost 400 years. The reasons for plague's persistence and abrupt disappearance in Europe are poorly understood, but could have been due to either the presence of now-extinct plague foci in Europe itself, or successive disease introductions from other locations. Here we present five Y. pestis genomes from one of the last European outbreaks of plague, from 1722 in Marseille, France. The lineage identified has not been found in any extant Y. pestis foci sampled to date, and has its ancestry in strains obtained from victims of the 14th century Black Death. These data suggest the existence of a previously uncharacterized historical plague focus that persisted for at least three centuries. We propose that this disease source may have been responsible for the many resurgences of plague in Europe following the Black Death.
Keywords: ancient DNA; epidemiology; global health; human history; infectious disease; microbial evolution; microbiology; pathogen genomics; yersinia pestis.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures

References
-
- Achtman M, Morelli G, Zhu P, Wirth T, Diehl I, Kusecek B, Vogler AJ, Wagner DM, Allender CJ, Easterday WR, Chenal-Francisque V, Worsham P, Thomson NR, Parkhill J, Lindler LE, Carniel E, Keim P. Microevolution and history of the plague bacillus, Yersinia pestis. Proceedings of the National Academy of Sciences. 2004;101:17837–17842. doi: 10.1073/pnas.0408026101. - DOI - PMC - PubMed
-
- Barnes KB. Social vulnerability and pneumonic plague: revisiting the 1994 outbreak in surat, india. Environmental Hazards. 2014;13:161–180. doi: 10.1080/17477891.2014.892867. - DOI
-
- Bos KI, Harkins KM, Herbig A, Coscolla M, Weber N, Comas I, Forrest SA, Bryant JM, Harris SR, Schuenemann VJ, Campbell TJ, Majander K, Wilbur AK, Guichon RA, Wolfe Steadman DL, Cook DC, Niemann S, Behr MA, Zumarraga M, Bastida R, Huson D, Nieselt K, Young D, Parkhill J, Buikstra JE, Gagneux S, Stone AC, Krause J. Pre-columbian mycobacterial genomes reveal seals as a source of new world human tuberculosis. Nature. 2014;514:494–497. doi: 10.1038/nature13591. - DOI - PMC - PubMed
-
- Bos KI, Schuenemann VJ, Golding GB, Burbano HA, Waglechner N, Coombes BK, McPhee JB, DeWitte SN, Meyer M, Schmedes S, Wood J, Earn DJD, Herring DA, Bauer P, Poinar HN, Krause J. A draft genome of yersinia pestis from victims of the black death. Nature. 2011;478:506–510. doi: 10.1038/nature10549. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

