Mucin glycan foraging in the human gut microbiome
- PMID: 25852737
- PMCID: PMC4365749
- DOI: 10.3389/fgene.2015.00081
Mucin glycan foraging in the human gut microbiome
Abstract
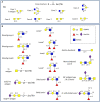
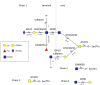
The availability of host and dietary carbohydrates in the gastrointestinal (GI) tract plays a key role in shaping the structure-function of the microbiota. In particular, some gut bacteria have the ability to forage on glycans provided by the mucus layer covering the GI tract. The O-glycan structures present in mucin are diverse and complex, consisting predominantly of core 1-4 mucin-type O-glycans containing α- and β- linked N-acetyl-galactosamine, galactose and N-acetyl-glucosamine. These core structures are further elongated and frequently modified by fucose and sialic acid sugar residues via α1,2/3/4 and α2,3/6 linkages, respectively. The ability to metabolize these mucin O-linked oligosaccharides is likely to be a key factor in determining which bacterial species colonize the mucosal surface. Due to their proximity to the immune system, mucin-degrading bacteria are in a prime location to influence the host response. However, despite the growing number of bacterial genome sequences available from mucin degraders, our knowledge on the structural requirements for mucin degradation by gut bacteria remains fragmented. This is largely due to the limited number of functionally characterized enzymes and the lack of studies correlating the specificity of these enzymes with the ability of the strain to degrade and utilize mucin and mucin glycans. This review focuses on recent findings unraveling the molecular strategies used by mucin-degrading bacteria to utilize host glycans, adapt to the mucosal environment, and influence human health.
Keywords: O-glycosylation; carbohydrate; gastrointestinal tract; glycoside hydrolase; gut bacteria; gut health and disease; intestinal mucus; mucin degradation.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

