Vertically transmitted faecal IgA levels determine extra-chromosomal phenotypic variation
- PMID: 25686606
- PMCID: PMC4425643
- DOI: 10.1038/nature14139
Vertically transmitted faecal IgA levels determine extra-chromosomal phenotypic variation
Abstract
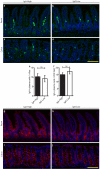
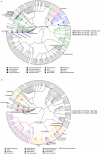
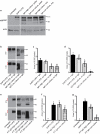
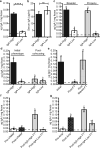
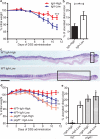
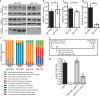
The proliferation of genetically modified mouse models has exposed phenotypic variation between investigators and institutions that has been challenging to control. In many cases, the microbiota is the presumed cause of the variation. Current solutions to account for phenotypic variability include littermate and maternal controls or defined microbial consortia in gnotobiotic mice. In conventionally raised mice, the microbiome is transmitted from the dam. Here we show that microbially driven dichotomous faecal immunoglobulin-A (IgA) levels in wild-type mice within the same facility mimic the effects of chromosomal mutations. We observe in multiple facilities that vertically transmissible bacteria in IgA-low mice dominantly lower faecal IgA levels in IgA-high mice after co-housing or faecal transplantation. In response to injury, IgA-low mice show increased damage that is transferable by faecal transplantation and driven by faecal IgA differences. We find that bacteria from IgA-low mice degrade the secretory component of secretory IgA as well as IgA itself. These data indicate that phenotypic comparisons between mice must take into account the non-chromosomal hereditary variation between different breeders. We propose faecal IgA as one marker of microbial variability and conclude that co-housing and/or faecal transplantation enables analysis of progeny from different dams.
Figures
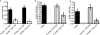



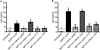
References
-
- Hao LY, Liu X, Franchi L. Inflammasomes in inflammatory bowel disease pathogenesis. Current opinion in gastroenterology. 2013;29:363–369. doi:10.1097/MOG.0b013e32836157a4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK097079/DK/NIDDK NIH HHS/United States
- P30DK052574/DK/NIDDK NIH HHS/United States
- R01 DK071619/DK/NIDDK NIH HHS/United States
- P30AR048335/AR/NIAMS NIH HHS/United States
- R01 DK101354/DK/NIDDK NIH HHS/United States
- T32AI007163/AI/NIAID NIH HHS/United States
- T32CA009547/CA/NCI NIH HHS/United States
- T32 CA009547/CA/NCI NIH HHS/United States
- DK7161907/DK/NIDDK NIH HHS/United States
- P30 AR048335/AR/NIAMS NIH HHS/United States
- T32 AI007163/AI/NIAID NIH HHS/United States
- AI08488702/AI/NIAID NIH HHS/United States
- R01 OD011170/OD/NIH HHS/United States
- P30 DK052574/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

