CHARMM-GUI Membrane Builder toward realistic biological membrane simulations
- PMID: 25130509
- PMCID: PMC4165794
- DOI: 10.1002/jcc.23702
CHARMM-GUI Membrane Builder toward realistic biological membrane simulations
Abstract
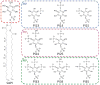
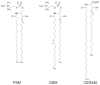
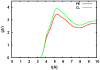
CHARMM-GUI Membrane Builder, http://www.charmm-gui.org/input/membrane, is a web-based user interface designed to interactively build all-atom protein/membrane or membrane-only systems for molecular dynamics simulations through an automated optimized process. In this work, we describe the new features and major improvements in Membrane Builder that allow users to robustly build realistic biological membrane systems, including (1) addition of new lipid types, such as phosphoinositides, cardiolipin (CL), sphingolipids, bacterial lipids, and ergosterol, yielding more than 180 lipid types, (2) enhanced building procedure for lipid packing around protein, (3) reliable algorithm to detect lipid tail penetration to ring structures and protein surface, (4) distance-based algorithm for faster initial ion displacement, (5) CHARMM inputs for P21 image transformation, and (6) NAMD equilibration and production inputs. The robustness of these new features is illustrated by building and simulating a membrane model of the polar and septal regions of E. coli membrane, which contains five lipid types: CL lipids with two types of acyl chains and phosphatidylethanolamine lipids with three types of acyl chains. It is our hope that CHARMM-GUI Membrane Builder becomes a useful tool for simulation studies to better understand the structure and dynamics of proteins and lipids in realistic biological membrane environments.
Keywords: cardiolipin; lipid penetration detection; phosphoinositides; sphingolipids.
Copyright © 2014 Wiley Periodicals, Inc.
Figures
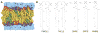
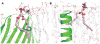
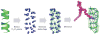
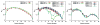
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

