Tackling soil diversity with the assembly of large, complex metagenomes
- PMID: 24632729
- PMCID: PMC3977251
- DOI: 10.1073/pnas.1402564111
Tackling soil diversity with the assembly of large, complex metagenomes
Erratum in
- Proc Natl Acad Sci U S A. 2014 Apr 22;111(16):6115
Abstract
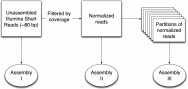
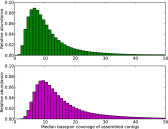
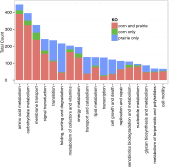
The large volumes of sequencing data required to sample deeply the microbial communities of complex environments pose new challenges to sequence analysis. De novo metagenomic assembly effectively reduces the total amount of data to be analyzed but requires substantial computational resources. We combine two preassembly filtering approaches--digital normalization and partitioning--to generate previously intractable large metagenome assemblies. Using a human-gut mock community dataset, we demonstrate that these methods result in assemblies nearly identical to assemblies from unprocessed data. We then assemble two large soil metagenomes totaling 398 billion bp (equivalent to 88,000 Escherichia coli genomes) from matched Iowa corn and native prairie soils. The resulting assembled contigs could be used to identify molecular interactions and reaction networks of known metabolic pathways using the Kyoto Encyclopedia of Genes and Genomes Orthology database. Nonetheless, more than 60% of predicted proteins in assemblies could not be annotated against known databases. Many of these unknown proteins were abundant in both corn and prairie soils, highlighting the benefits of assembly for the discovery and characterization of novelty in soil biodiversity. Moreover, 80% of the sequencing data could not be assembled because of low coverage, suggesting that considerably more sequencing data are needed to characterize the functional content of soil.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Hess M, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331(6016):463–467. - PubMed
-
- Iverson V, et al. Untangling genomes from metagenomes: Revealing an uncultured class of marine Euryarchaeota. Science. 2012;335(6068):587–590. - PubMed
-
- Mackelprang R, et al. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature. 2011;480(7377):368–371. - PubMed

