Significant changes in the skin microbiome mediated by the sport of roller derby
- PMID: 23638391
- PMCID: PMC3628844
- DOI: 10.7717/peerj.53
Significant changes in the skin microbiome mediated by the sport of roller derby
Abstract
Diverse bacterial communities live on and in human skin. These complex communities vary by skin location on the body, over time, between individuals, and between geographic regions. Culture-based studies have shown that human to human and human to surface contact mediates the dispersal of pathogens, yet little is currently known about the drivers of bacterial community assembly patterns on human skin. We hypothesized that participation in a sport involving skin to skin contact would result in detectable shifts in skin bacterial community composition. We conducted a study during a flat track roller derby tournament, and found that teammates shared distinct skin microbial communities before and after playing against another team, but that opposing teams' bacterial communities converged during the course of a roller derby bout. Our results are consistent with the hypothesis that the human skin microbiome shifts in composition during activities involving human to human contact, and that contact sports provide an ideal setting in which to evaluate dispersal of microorganisms between people.
Keywords: Contact sport; Human microbiome; Microbial biogeography; Microbial dispersal; Microbial ecology; Skin microbiology.
Figures
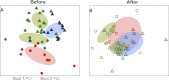
 vs. SI
vs. SI  ) and before bout 2 (EC
) and before bout 2 (EC  vs. DC
vs. DC  ). Corresponding-colored ellipses show standard deviations around community variances from each team. The skin bacterial communities of the four team groups were significantly different before playing a bout (p < 0.001; from permutational MANOVA on Canberra taxonomic distances). (B) The four team groups are also significantly different after playing bouts (p < 0.001), though more overlap is observed between teams after bout 1 (EC
). Corresponding-colored ellipses show standard deviations around community variances from each team. The skin bacterial communities of the four team groups were significantly different before playing a bout (p < 0.001; from permutational MANOVA on Canberra taxonomic distances). (B) The four team groups are also significantly different after playing bouts (p < 0.001), though more overlap is observed between teams after bout 1 (EC  vs. SI
vs. SI  ) and after bout 2 (EC
) and after bout 2 (EC  vs. DC
vs. DC  ). NMDS 3-dimensional stress = 19.66 (A) & 17.55 (B).
). NMDS 3-dimensional stress = 19.66 (A) & 17.55 (B).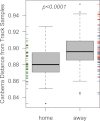

 and after
and after  bout 1; (B) Silicon Valley before
bout 1; (B) Silicon Valley before  and after
and after  bout 1; (C) Emerald City before
bout 1; (C) Emerald City before  and after
and after  bout 2; (D) DC before
bout 2; (D) DC before  and after
and after  bout 2. Corresponding-colored ellipses are standard deviations on community variances for each group. All teams showed significantly different microbial communities before vs. after a bout. NMDS 3-dimensional stress: A = 8.1, B = 10.47, C = 16.2, D = 17.65.
bout 2. Corresponding-colored ellipses are standard deviations on community variances for each group. All teams showed significantly different microbial communities before vs. after a bout. NMDS 3-dimensional stress: A = 8.1, B = 10.47, C = 16.2, D = 17.65.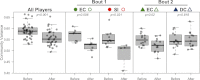
References
-
- Blaser MJ, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Estrada I, Gao Z, Clemente JC, Costello EK, Knight R. Distinct cutaneous bacterial assemblages in a sampling of south american amerindians and us residents. The ISME Journal: Multidisciplinary Journal of Microbial Ecology. 2012;7:85–95. doi: 10.1038/ismej.2012.81. - DOI - PMC - PubMed
-
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

