Plant intracellular innate immune receptor Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane
- PMID: 21490299
- PMCID: PMC3088580
- DOI: 10.1073/pnas.1104410108
Plant intracellular innate immune receptor Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane
Erratum in
- Proc Natl Acad Sci U S A. 2011 May 24;108(21):8915. Gao, Zhiyoug [corrected to Gao, Zhiyong]
Abstract
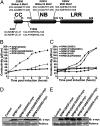
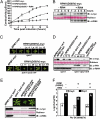
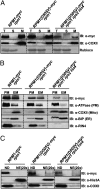
Plants deploy intracellular innate immune receptors to recognize pathogens and initiate disease resistance. These nucleotide-binding, leucine-rich repeat (NB-LRR) proteins are activated by pathogen effector proteins that are delivered into the host cell to suppress host defense responses. Little is known about the sites and mechanisms of NB-LRR activation, but some NB-LRR proteins can function inside the plant nucleus. We demonstrate that RPM1 is activated on the plasma membrane and does not relocalize to the nucleus. An autoactive RPM1(D505V) allele that recapitulates key features of normal RPM1 activation also resides on the plasma membrane. There is no detectable relocalization of activated RPM1 to the nucleus. Hindering potential nuclear entry of RPM1-Myc did not affect either its effector-triggered hypersensitive-response (HR) cell death or its disease resistance functions, further suggesting that nuclear translocation is not required for RPM1 function. RPM1 tethered onto the plasma membrane with a dual acylated N-terminal epitope tag retained the ability to mediate HR, consistent with this RPM1 function being activated on the plasma membrane. Plant NB-LRR proteins can thus function at various locations in the cell.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. - PubMed
-
- Dodds PN, Rathjen JP. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–548. - PubMed
-
- Ting JP, Willingham SB, Bergstralh DT. NLRs at the intersection of cell death and immunity. Nat Rev Immunol. 2008;8:372–379. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

