A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci
- PMID: 18369459
- PMCID: PMC2274885
- DOI: 10.1371/journal.pgen.1000041
A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci
Abstract
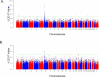
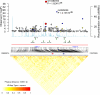
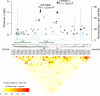
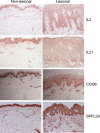
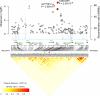
A genome-wide association study was performed to identify genetic factors involved in susceptibility to psoriasis (PS) and psoriatic arthritis (PSA), inflammatory diseases of the skin and joints in humans. 223 PS cases (including 91 with PSA) were genotyped with 311,398 single nucleotide polymorphisms (SNPs), and results were compared with those from 519 Northern European controls. Replications were performed with an independent cohort of 577 PS cases and 737 controls from the U.S., and 576 PSA patients and 480 controls from the U.K.. Strongest associations were with the class I region of the major histocompatibility complex (MHC). The most highly associated SNP was rs10484554, which lies 34.7 kb upstream from HLA-C (P = 7.8x10(-11), GWA scan; P = 1.8x10(-30), replication; P = 1.8x10(-39), combined; U.K. PSA: P = 6.9x10(-11)). However, rs2395029 encoding the G2V polymorphism within the class I gene HCP5 (combined P = 2.13x10(-26) in U.S. cases) yielded the highest ORs with both PS and PSA (4.1 and 3.2 respectively). This variant is associated with low viral set point following HIV infection and its effect is independent of rs10484554. We replicated the previously reported association with interleukin 23 receptor and interleukin 12B (IL12B) polymorphisms in PS and PSA cohorts (IL23R: rs11209026, U.S. PS, P = 1.4x10(-4); U.K. PSA: P = 8.0x10(-4); IL12B:rs6887695, U.S. PS, P = 5x10(-5) and U.K. PSA, P = 1.3x10(-3)) and detected an independent association in the IL23R region with a SNP 4 kb upstream from IL12RB2 (P = 0.001). Novel associations replicated in the U.S. PS cohort included the region harboring lipoma HMGIC fusion partner (LHFP) and conserved oligomeric golgi complex component 6 (COG6) genes on chromosome 13q13 (combined P = 2x10(-6) for rs7993214; OR = 0.71), the late cornified envelope gene cluster (LCE) from the Epidermal Differentiation Complex (PSORS4) (combined P = 6.2x10(-5) for rs6701216; OR 1.45) and a region of LD at 15q21 (combined P = 2.9x10(-5) for rs3803369; OR = 1.43). This region is of interest because it harbors ubiquitin-specific protease-8 whose processed pseudogene lies upstream from HLA-C. This region of 15q21 also harbors the gene for SPPL2A (signal peptide peptidase like 2a) which activates tumor necrosis factor alpha by cleavage, triggering the expression of IL12 in human dendritic cells. We also identified a novel PSA (and potentially PS) locus on chromosome 4q27. This region harbors the interleukin 2 (IL2) and interleukin 21 (IL21) genes and was recently shown to be associated with four autoimmune diseases (Celiac disease, Type 1 diabetes, Grave's disease and Rheumatoid Arthritis).
Conflict of interest statement
Patent application on findings in process.
Figures

Comment in
-
Genome-wide association scan in psoriasis: new insights into chronic inflammatory disease.Expert Rev Clin Immunol. 2008 Sep;4(5):565-71. doi: 10.1586/1744666X.4.5.565. Expert Rev Clin Immunol. 2008. PMID: 20476959
References
-
- Bowcock AM, Krueger JG. Getting under the skin: The immunogenetics of psoriasis. Nat Rev Immunol. 2005;5:699–711. - PubMed
-
- Fitzgerald O, Dougados M. Psoriatic arthritis: one or more diseases? Best Practice & Research in Clinical Rheumatology. 2006;20:435–450. - PubMed
-
- Ritchlin CT. Pathogenesis of psoriatic arthritis. Curr Op Rheum. 2005;17:406–412. - PubMed
-
- Gladman DD, Farewell VT, Pellett F, Schentag C, Rahman P. HLA is a candidate region for psoriatic arthritis. evidence for excessive HLA sharing in sibling pairs. Hum Immunol. 2003;64:887–889. - PubMed
-
- Myers A, Kay LJ, Lynch SA, Walker DJ. Recurrence risk for psoriasis and psoriatic arthritis within sibships. Rheumatology (Oxford) 2005;44:773–776. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

