eQED: an efficient method for interpreting eQTL associations using protein networks
- PMID: 18319721
- PMCID: PMC2290938
- DOI: 10.1038/msb.2008.4
eQED: an efficient method for interpreting eQTL associations using protein networks
Abstract
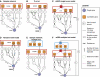
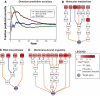
Analysis of expression quantitative trait loci (eQTLs) is an emerging technique in which individuals are genotyped across a panel of genetic markers and, simultaneously, phenotyped using DNA microarrays. Because of the spacing of markers and linkage disequilibrium, each marker may be near many genes making it difficult to finely map which of these genes are the causal factors responsible for the observed changes in the downstream expression. To address this challenge, we present an efficient method for prioritizing candidate genes at a locus. This approach, called 'eQTL electrical diagrams' (eQED), integrates eQTLs with protein interaction networks by modeling the two data sets as a wiring diagram of current sources and resistors. eQED achieved a 79% accuracy in recovering a reference set of regulator-target pairs in yeast, which is significantly higher than the performance of three competing methods. eQED also annotates 368 protein-protein interactions with their directionality of information flow with an accuracy of approximately 75%.
Figures
References
-
- Albert-László Barabási RA (1999) Emergence of scaling in random networks. Science 286: 509–512 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

