Parkin occurs in a stable, non-covalent, approximately 110-kDa complex in brain
- PMID: 18190519
- PMCID: PMC2253705
- DOI: 10.1111/j.1460-9568.2007.06000.x
Parkin occurs in a stable, non-covalent, approximately 110-kDa complex in brain
Abstract
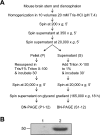
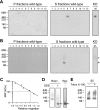
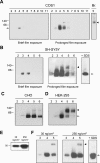
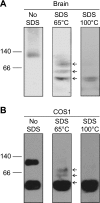
Mutations in the gene for parkin, a 52-kDa E3 ubiquitin ligase, are a major cause of hereditary Parkinson's disease (PD). In vitro studies have identified a large number of parkin-interacting proteins. Whether parkin exists as a monomer or as part of a stable protein complex in vivo is uncertain. Here we demonstrate that endogenous parkin occurs in a stable, non-covalent, approximately 110-kDa complex in native extracts from mouse brain, heart and skeletal muscle, while monomeric parkin is undetectable. Partial denaturation experiments indicate that this complex is at least a tetramer. Reported parkin-binding partners do not show detectable association with the parkin complex on native gels. Upon overexpression in COS1, SH-SY5Y or CHO cells, parkin accumulates predominantly as a monomer, suggesting that the interactors required for complex formation are available in limiting amounts in these cells. Importantly, PD-linked parkin mutations significantly impair parkin complex formation. These data demonstrate that parkin oligomerizes into a stable, non-covalent, heteromeric complex in vivo, and suggest that parkin may have as yet unidentified stable binding partners.
Figures




References
-
- Baulac S, Lavoie MJ, Strahle J, Schlossmacher MG, Xia W. Dimerization of Parkinson's disease-causing DJ-1 and formation of high molecular weight complexes in human brain. Mol. Cell. Neurosci. 2004;27:236–246. - PubMed
-
- Capell A, Grunberg J, Pesold B, Diehlmann A, Citron M, Nixon R, Beyreuther K, Selkoe DJ, Haass C. The proteolytic fragments of the Alzheimer's disease-associated presenilin-1 form heterodimers and occur as a 100–150-kDa molecular mass complex. J. Biol. Chem. 1998;273:3205–3211. - PubMed
-
- Chung KK, Zhang Y, Lim KL, Tanaka Y, Huang H, Gao J, Ross CA, Dawson VL, Dawson TM. Parkin ubiquitinates the α-synuclein-interacting protein, synphilin-1: implications for Lewy-body formation in Parkinson disease. Nat. Med. 2001;7:1144–1150. - PubMed
-
- Corti O, Hampe C, Koutnikova H, Darios F, Jacquier S, Prigent A, Robinson JC, Pradier L, Ruberg M, Mirande M, Hirsch E, Rooney T, Fournier A, Brice A. The p38 subunit of the aminoacyl-tRNA synthetase complex is a parkin substrate: linking protein biosynthesis and neurodegeneration. Hum. Mol. Genet. 2003;15:1427–1437. - PubMed
-
- Dachsel JC, Lücking CB, Deeg S, Schultz E, Lalowski M, Casademunt E, Corti O, Hampe C, Patenge N, Vaupel K, Yamamoto A, Dichgans M, Brice A, Wanker EE, Kahle PJ, Gasser T. Parkin interacts with the proteasome subunit α4. FEBS Lett. 2005;579:3913–3919. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

