Increased glycolytic flux as an outcome of whole-genome duplication in yeast
- PMID: 17667951
- PMCID: PMC1943425
- DOI: 10.1038/msb4100170
Increased glycolytic flux as an outcome of whole-genome duplication in yeast
Erratum in
- Mol Syst Biol. 2008;4():204
Abstract
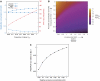
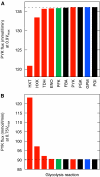
After whole-genome duplication (WGD), deletions return most loci to single copy. However, duplicate loci may survive through selection for increased dosage. Here, we show how the WGD increased copy number of some glycolytic genes could have conferred an almost immediate selective advantage to an ancestor of Saccharomyces cerevisiae, providing a rationale for the success of the WGD. We propose that the loss of other redundant genes throughout the genome resulted in incremental dosage increases for the surviving duplicated glycolytic genes. This increase gave post-WGD yeasts a growth advantage through rapid glucose fermentation; one of this lineage's many adaptations to glucose-rich environments. Our hypothesis is supported by data from enzyme kinetics and comparative genomics. Because changes in gene dosage follow directly from post-WGD deletions, dosage selection can confer an almost instantaneous benefit after WGD, unlike neofunctionalization or subfunctionalization, which require specific mutations. We also show theoretically that increased fermentative capacity is of greatest advantage when glucose resources are both large and dense, an observation potentially related to the appearance of angiosperms around the time of WGD.
Figures

References
-
- Aon MA, Mónaco ME, Cortassa S (1995) Carbon and energetic uncoupling are associated with block of division at different stages of the cell cycle in several cdc mutants of Saccharomyces cerevisiae. Exp Cell Res 217: 42–51 - PubMed
-
- Ashburner A (1998) Speculations on the subject of alcohol dehydrogenase and its properties in Drosophila and other flies. BioEssays 20: 949–954 - PubMed
-
- Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurens B, Daubin V, Anthouard V, Aiach N, Arnaiz O, Billaut A, Beisson J, Blanc I, Bouhouche K, Camara F, Duharcourt S, Guigo R, Gogendeau D, Katinka M, Keller AM, Kissmehl R, Klotz C, Koll F, Le Mouel A, Lepere G, Malinsky S, Nowacki M, Nowak JK, Plattner H, Poulain J, Ruiz F, Serrano V, Zagulski M, Dessen P, Betermier M, Weissenbach J, Scarpelli C, Schachter V, Sperling L, Meyer E, Cohen J, Wincker P (2006) Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444: 171–178 - PubMed
-
- Bakalinsky AT, Snow R (1990) The chromosomal constitution of wine strains of Saccharomyces cerevisiae. Yeast 6: 367–382 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

