Effect of temperature on geminivirus-induced RNA silencing in plants
- PMID: 16040661
- PMCID: PMC1183375
- DOI: 10.1104/pp.105.066563
Effect of temperature on geminivirus-induced RNA silencing in plants
Abstract
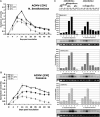
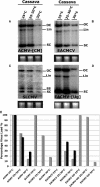
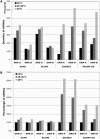
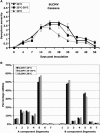
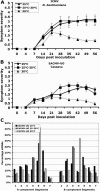
Short-interfering RNAs (siRNAs), the molecular markers of posttranscriptional gene silencing (PTGS), are powerful tools that interfere with gene expression and counter virus infection both in plants and animals. Here, we report the effect of temperature on geminivirus-induced gene silencing by quantifying virus-derived siRNAs and by evaluating their distribution along the virus genome for isolates of five species of cassava geminiviruses in cassava (Manihot esculenta, Crantz) and Nicotiana benthamiana. Cassava geminivirus-induced RNA silencing increased by raising the temperature from 25 degrees C to 30 degrees C, with the appearance of less symptomatic newly developed leaves, irrespective of the nature of the virus. Consequently, nonrecovery-type geminiviruses behaved like recovery-type viruses under high temperature. Next, we evaluated the distribution of virus-derived siRNAs on the respective virus genome at three temperatures (25 degrees C, 25 degrees C-30 degrees C, and 30 degrees C). For recovery-type viruses, siRNAs accumulated at moderately higher levels during virus-induced PTGS at higher temperatures, and there was no change in the distribution of the siRNA population along the virus genome. For nonrecovery-type viruses, siRNAs accumulated at strikingly higher levels than those observed for infections with recovery-type viruses at high temperature. As determined for an RNA virus, temperature influences gene silencing for single-stranded DNA geminiviruses. It is possible that other mechanisms besides gene silencing also control geminivirus accumulation at high temperatures. The findings presented here should be taken into consideration when implementing PTGS-based strategies to control plant virus accumulation.
Figures


References
-
- Baulcombe D (2004) RNA silencing in plants. Nature 431: 356–363 - PubMed
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 - PubMed
-
- Chellappan P, Masona MV, Vanitharani R, Taylor NJ, Fauquet CM (2004. b) Broad spectrum resistance to ssDNA viruses associated with transgene-induced gene silencing in cassava. Plant Mol Biol 56: 601–611 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

