Abstract
Candida auris is a newly emerging species that was first identified in Asia in 2009 but has rapidly spread across the world. C. auris differs from most other Candida species in that antifungal resistance is the norm rather than the exception, it is a commensal of human skin rather than the human gut, and it can be easily transmitted from person to person in a healthcare setting. This review discusses the emergence of C. auris, global epidemiology, identification, antifungal susceptibility testing, and precautions to be taken when it is identified from a patient specimen.
Introduction
“Have you heard about this new Candida species, Candida auris?”
If you haven’t gotten that question yet, chances are you soon will. Candida auris was first described as a species in 2009 but it is still a relatively new to clinical microbiology laboratories [1]. The first described isolate was from the ear canal of a patient in Japan, but C. auris soon started showing up as a cause of bloodstream infection in other parts of Asia including Korea, India, and Kuwait [2–5]. Over the past eight years it has slowly started to emerge on other continents; first in Africa, then South America, and most recently in North America and Europe [6–12]. C. auris is closely related to three other rare Candida species, C. haemulonii, C. duobushaemulonii, and C. pseudohaemulonii in the family Metschnikowiaceae along with the more common Candida lusitaniae [13]. Although very few members of this family are known to cause human infection, those that do are known for their ability to develop resistance to fluconazole and amphotericin B. As will be mentioned below, C. auris can be difficult to distinguish from isolates of the C. haemulonii species complex in the clinical microbiology laboratory.
Where in the world did C. auris originate?
Although the first reports of C. auris came out of Asia, shortly after there were subsequent reports of isolates from South Africa as well as Venezuela [6, 7]. One of the major questions surrounding C. auris was how it was able to emerge in so many countries in such a short time when it was essentially unheard of prior to the first report in 2009; did it emerge independently in each country or did a single outbreak strain spread from an original source? Whole-genome sequencing (WGS) and single-nucleotide polymorphism (SNP) analysis allow the analysis of C. auris isolates in high resolution, by characterizing genetic diversity of the 12.5 million base pairs in the genomes, which helps to differentiate even the most closely related isolates. C. auris reference genomes were assembled using two different types of whole genome sequencing systems, Illumina and PacBio, and those sequences were published and made publically available [9, 14, 15]. Using these methods and resources, phylogenetic trees that reveal population dynamics and structure were constructed, so that when coupled with the epidemiology, a better understanding of the spread of C. auris could be obtained.
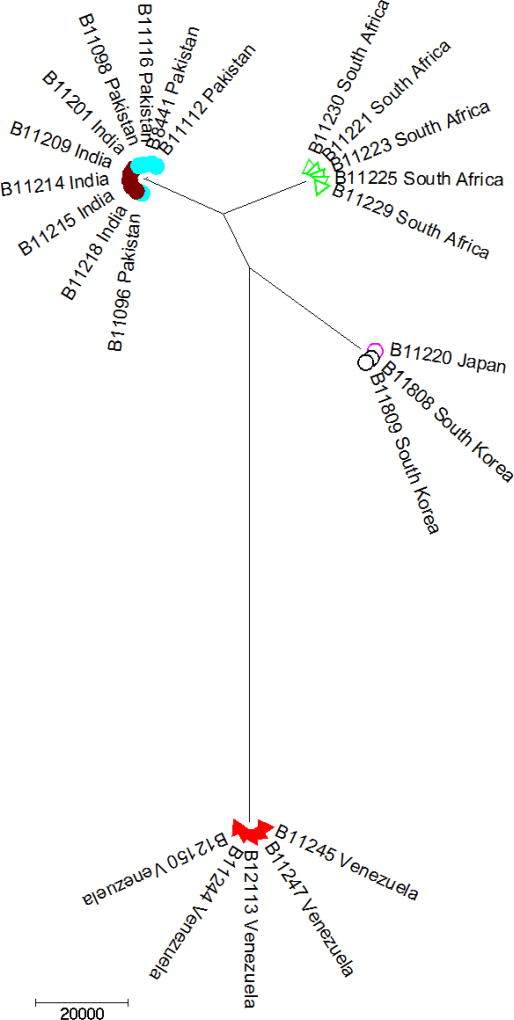
In a study that was initiated by the Centers for Disease Control and Prevention, C. auris isolates from Japan, India, Pakistan, South Africa, and Venezuela were sequenced and the results were startling [9]. Supporting previous studies that employed more traditional molecular and proteomic methods, isolates were found to cluster by geography [2, 3, 16] (Figure 1). The remarkable finding was that isolates grouped to clusters that were nearly identical within the cluster, but the clusters themselves were genetically unrelated to each other. Specifically, four clades characterized by region (East Asian, South Asian, African, and South American) were identified. Each clade was separated by 40,000 – 140,000 single nucleotide polymorphisms (or SNPs). Within each clade, the average number of SNP differences between any two isolates was <70 (with many being <10), indicating a high degree of clonality within clades. Together, these results suggest an independent, nearly simultaneous emergence of four populations on three continents. Why and how this occurred remains unknown. Multiple independent laboratories with international culture collections subsequently reviewed older isolates to see if C. auris had been isolated previously and was either misidentified or not identified at all. While a single isolate from 1996 in Korea had been misidentified and a single isolate from Pakistan in 2008 had been unidentified, no other isolates of C. auris were identified from over 30,000 isolates from more than 40 countries that were reviewed [4, 9] (CDC, unpublished data). This corroborates the recent clinical emergence of C. auris within the last 10 years.
Figure 1.
Neighbor-joining dendrogram of whole genome sequences of Candida auris. Each color represents a unique country. The isolates fall into four very distinct clusters that differ widely from each other but are internally very clonal.
Combining phylogenetic analysis with epidemiology can help public health officials track the spread of C. auris. This has been particularly true in the United States. As of August 31, 2016, seven U.S. cases of C. auris infection were reported to the CDC [10]. Cases were reported from four states; two from a single hospital in Illinois, one from Maryland, one from New Jersey, and three from three different hospitals in New York. WGS was performed on isolates from six cases, and surprisingly, isolates did not form a new clade distinct from the four known clades. Instead, isolates from the east coast of the United States (Maryland, New Jersey, and New York) clustered with the South Asian clade with a pairwise difference of around 60 SNPs when compared to the Indian and Pakistani isolates. As for the cases from Illinois, isolates clustered with the South American clade, differing by around 150 SNPs from the Venezuelan isolates. Although none of the cases from which isolates were sequenced had known travel or other direct links to South Asia or South America, the data suggests an original transmission link between these regions and the United States. Since the time of that report, the case count in the United States has increased to 33 (https://www.cdc.gov/fungal/diseases/candidiasis/candida-auris.html; accessed 2/15/2017). However, all of the additional cases have been identified either in Illinois or New York, and each can be linked epidemiologically to a previous case. The majority of the cases of C. auris that have been reported either show multiple cases in a single institution, or report the clonal relatedness of isolates from multiple institutions [2, 3, 7–9, 11, 12, 17]. This suggests that there is transmission within the healthcare setting. This is unusual for Candida. Although outbreaks of Candida do occasionally occur, it is thought that the majority of Candida infections are endogenous; patients are infected with their own commensal flora. However, data from patient and environmental isolates suggests that transmission of C. auris occurs within healthcare settings [12]. During an outbreak at a hospital in England, patients were colonized with C. auris in multiple body sites including axilla, groin, nares, ear, rectum, and in urine. With the exception of C. parapsilosis, Candida is generally thought of as gut flora and not a skin colonizer. In addition, C. auris can be detected in the environment of the room of a colonized or infected patient, including on the bed, windowsills, tables equipment monitors, and the floor [10, 12], more like what would be expected for a bacteria like Clostridium difficile or an Acinetobacter and not a fungus.
The epidemiological indicators of healthcare-associated transmission are corroborated by the WGS data. Fewer SNP differences exist between cases who have shared a healthcare facility as compared to those between cases who have not shared a common facility. In the case of the seven reported U.S. cases, both the Maryland and New Jersey cases were at one point admitted to the same New Jersey hospital at the same time. These isolates differed by <10 SNPs, which was comparable to the difference shared between the two Illinois cases that were admitted to the same hospital (<10 SNPs), to the difference between environmental and case isolates from the Illinois hospital (<5 SNPs), and also to the difference between multiple isolates sequenced from a single patient over a 10-day period (<6 SNPs) [10]. Although not as stringent as WGS, amplified fragment length polymorphism (AFLP) typing also indicates that isolates identified within an institution are more closely related to each other than isolates from other institutions [2, 3, 7, 12]
Are we going to be able to identify it in our laboratory?
Although the data presented above indicates that it has truly emerged only recently, Candida auris has likely been under reported due to unreliable identification [4, 18, 19]. Rare pathogens are often misidentified simply for the fact that they are not included in the databases of commercially available rapid diagnostic systems, and this has certainly been the case with C. auris. Unfortunately, this can delay accurate detection by clinical laboratories and treatment by clinicians, further compounding the underreporting in the field. To allow laboratories to validate their own internal identification systems and to allow commercial companies to get it into their databases, the CDC has recently made a panel of C. auris isolates and closely-related species publicly available through the FDA/CDC AR Bank (https://www.cdc.gov/drugresistance/resistance-bank/form/index.html).
Standard biochemical identification systems are unreliable for identifying C. auris primarily because they do not have C. auris in their databases. C. auris is misidentified as Candida haemulonii by the Vitek-2 (bioMérieux), and as Rhodotorula glutinis, Candida sake or Saccharomyces cerevisiae by the API 20C AUX (bioMérieux) systems [4, 9, 18]. The CDC panel of ten C. auris isolates with representatives from each of the four clades (as described above) were misidentified as C. haemulonii, except for one as C. catenulata, by BD Phoenix (BD Diagnostics, Sparks, MD) and they were misidentified as C. parapsilosis, C. famata, C. lusitaniae, and C. guillermondii by MicroScan (Beckman Coutler, Pasadena, CA) [18]. Users of these biochemical identification systems should realize that they may not be able to identify C. auris or possibly even rule it out from some of the more common Candida species. As these systems update their databases, accurate identifications may become possible. For example, bioMerieux has reported that their most recent software update for the VITEK 2 YST card (with Ver 8.01 software), which is just beginning to be released, allows the closely related species, C. auris, C. duobushaemulonii, and C. haemulonii to be correctly identified.
Sequencing of the internal transcribed spacer or the D1/D2 region of the ribosomal DNA provides accurate species level identification, but the cost, technical demands, and lengthy turnaround times for those without in-house sequencing capacity makes this less suitable for some laboratories. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) is a broad spectrum tool capable of faster and more accurate identification of most bacterial and fungal clinical isolates. MALDI-TOF takes a microbial sample input and produces a unique protein profile spectrum capable of discriminating microbes to the genus and species level [20]. The spectral profile is unique to the organisms being analyzed, and an accurate phylogenetic classification of the organism is automatically identified by the instrument using software to compare the spectral profile to reference databases. However, the important limitation to MALDI-TOF MS is that it can only identify organisms that are in the reference databases. A recent study comparing the Vitek-MS (bioMérieux) and the Bruker Biotyper (Billerica, MA) to their respective FDA-approved libraries using the panel of ten CDC C. auris isolates found that neither instrument were able to identify C. auris because it was not included in the database [18]. Identification is possible after installing the research-use-only Saramis Version 4.14 database and Saccharomycetaceae update for Vitek-MS or Bruker’s 6903 MSP databases for Biotyper [19, 21]. CDC’s MicrobeNet (https://www.cdc.gov/microbenet/index.html) is a unique tool designed to provide subject matter expert-curated information for the most relevant identification methods in today’s laboratories, including MALDI-TOF. In collaboration with Bruker, MicrobeNet recently released the Biotyper Classification Module which now provides MicrobeNet users with access to CDC spectral libraries as well as Bruker’s most up-to-date database. The strains of C. auris represented in the MicrobeNet database represent all of the known worldwide clades and accurately classify to the species level on the Biotyper.
The time until the initiation of antimicrobial treatment impacts patient mortality, so rapid and accurate diagnostic methods are a critical component of patient care. Historically we have endured lengthy turnaround times with clinical laboratories running phenotype testing and conventional identification methods. As most rapid candidemia treatment is empiric, the slower time to species identification has not been a problem. However, in the case of C. auris this does pose a problem because of the infection control considerations and the need to implement contact precautions (As will be outlined below). Infection prevention guided by rapid accurate detection in healthcare settings is essential for containment of this pathogen. MALDI-TOF MS has greatly decreased the turnaround, and it has seen rapid adoption worldwide, quickly becoming the standard practice in clinical microbiology [22, 23]. The limitations of the other identification systems, particularly with the available databases, should be closely monitored by lab directors, clinicians, and technologists, especially when it comes to emerging pathogens, because there is often a lag before these organisms are added to the various databases.
What about susceptibility testing, most Candida is antifungal susceptible, right?
The number of antifungal compounds that are available is very limited and essentially consists of only three classes; azoles, echinocandins, and polyenes. With this limitation in place, identifying species capable of developing resistance to those compounds is important in shaping treatment plans. Susceptibility is measured clinically as the reduction in growth in vitro as compared to drug-free control growth. This yields the minimum inhibitory concentration, or MIC, value which is used as a measure of antifungal activity. Broadly, Candida species are quite susceptible to antifungal drugs but exceptions include intrinsic fluconazole resistance in C. krusei and acquired echinocandin resistance in C. glabrata [24]. Approximately 6–8% of all clinical Candida isolates are resistant to fluconazole while 1% are resistant to the echinocandins, the majority in both categories being C. glabrata [25]. Amphotericin B resistance is extremely rare in Candida species with the exception of C. lusitaniae [26].
That being said, susceptibility testing for an emerging fungal species is complicated by the fact that little susceptibility information has been compiled, there are usually no breakpoints, and the correlation, if any, between MIC values and clinical outcome is not yet known. This is the case for C. auris. But by using breakpoints for other Candida species as guidance tentative MIC values defining resistance have been proposed (https://www.cdc.gov/fungal/diseases/candidiasis/recommendations.html). The MIC values for a very large number of isolates have been determined, and the distribution contains isolates on both the higher and lower end of the tested range. The isolates on the lower end of the range are used to set the values for susceptible isolates while those on the higher end help determine the MIC at which an isolate may be considered resistant. While not an official breakpoint, this serves as a guideline to use with such an emerging species. What has been determined regarding C. auris and antifungal susceptibility can be summed up in three words: it is concerning.
This species can be highly resistant to fluconazole (MIC ≥32 µg/mL), and can also show cross resistance to voriconazole (MIC ≥4 µg/mL) [3, 8–10, 17]. This is troubling given that the azoles are a mainstay in treatment of Candida infections and access to antifungals other than fluconazole is often poor in resource-limited countries. Some initial reports had nearly universal fluconazole resistance and the species was initially thought to have intrinsic resistance [3, 19]. However, as more isolates from around the world have been tested, isolates with fluconazole MIC values similar to those of C. glabrata (in the 2–8 µg/mL range) have been collected and fluconazole resistance is now thought to be acquired (CDC, unpublished data). In a recent study, 54 isolates of C. auris were analyzed for their antifungal susceptibilities and 93% and 33% were resistant to fluconazole and voriconazole, respectively [9]. In this study it was determined that the majority of the isolates with elevated MIC values to fluconazole and voriconazole carried mutations in the protein target of the azoles that had been previously shown to impact drug resistance in other Candida species. These mutations were geographically-distinct, with the isolates within a particular region having the same mutation, but different mutations for each region. Those isolates that did not carry such mutations had much lower MIC values to the azole drug class, suggesting that these isolates carried the wildtype version of this protein and were most likely susceptible. All reported MICs for both itraconazole and posaconazole have been relatively low; there are no isolates with elevated MICs which might be considered resistant. Because the chemical structure of intraconazole and posaconazole are somewhat different from fluconazole and voriconazole, it is not known whether this due to the shape of the target molecule or whether the isolates have just never been exposed to those two compounds which are rarely used to treat candidemia.
There are some isolates that exhibit resistance to one or more echinocandin (MIC ≥4 µg/mL for anidulafungin and micafungin, MIC ≥2 µg/mL for caspofungin) and even some with resistance to amphotericin B (MIC >2 µg/mL). An analysis of 102 C. auris isolates from 4 hospitals in India reported isolates to have a 37% resistance rate to caspofungin by broth microdilution [19]. In fact, other reports describe several isolates from various countries with resistance to one or more echinocandins or with resistance to amphotericin B [7, 9, 12] To date, the mechanisms underlying these types of resistance are unclear. Even more concerning are those isolates which show in vitro resistance to all 3 major classes of antifungals [9]. This leaves no further option for treatment and will pose an important clinical challenge if C. auris increases in prevalence.
It is worth reiterating that breakpoints for C. auris are not established and what is considered resistant at this point should be taken as a general rule and not definitive. Clinicians should not necessarily rule out the use of a specific antifungal based on MIC findings; treatment decisions should be made on a case-by-case basis. However, the high occurrence of in vitro antifungal resistance among clinical isolates of C. auris is unlike what has been observed in other species of Candida and is a serious concern.
Is there anything else we need to consider when dealing with C. auris?
As the title of the article suggests, C. auris is not the typical Candida species. As outlined above, it seems to be a skin commensal rather than gut flora, development of antifungal resistance seems to be the norm, and unlike the majority of Candida species, it can extensively contaminate healthcare environments, a scenario more typical of bacteria than fungi. For these considerations, the Centers for Disease Control and Prevention, and Public Health England each released clinical alerts, followed shortly by a Rapid risk assessment released by the European Centers for Disease Control (https://www.cdc.gov/fungal/diseases/candidiasis/candida-auris-alert.html; https://www.gov.uk/government/publications/candida-auris-laboratory-investigation-management-and-infection-prevention-and-control; http://ecdc.europa.eu/en/publications/Publications/Candida-in-healthcare-settings_19-Dec-2016.pdf). Each of these publications provides guidance to both clinicians and clinical laboratories on how to identify and report C. auris as well as infection control practices and environmental cleaning. The CDC recommendations are updated on a continuing basis and can be found here: https://www.cdc.gov/fungal/diseases/candidiasis/recommendations.html. Aside from what has been outlined above, the CDC recommends that patients who are infected or colonized by C. auris be placed under contact precautions to prevent the spread to other patients and to the hospital environment. This has never been a recommendation for any other Candida species. In addition, there is now a recommendation for daily and terminal cleaning with an EPA-registered hospital-grade disinfectant effective against Clostridium difficile spores. C. auris is not a nationally notifiable disease. However, CDC encourages all U.S. laboratory staff who identify C. auris strains to notify their state or local public health authorities and CDC at candidaauriscdc.gov.
Summary
Candida auris is the fungal equivalent of the new kid on the block. Our knowledge of this emerging organism is at the point where what we know about C. auris is less than what we do not know about C. auris. There are many important questions that remain to be answered. Why has it suddenly emerged? What was its prior natural niche? Will it continue to spread across the world? Can we decolonize people once they are colonized? With the assistance from clinical microbiology laboratories in the US and throughout the world, public health and laboratories, infection control specialists, and clinicians can begin to answer these questions. The first step is awareness of the problem at hand and it is hoped that this short review helped further your awareness and understanding of the emerging threat that is C. auris.
Footnotes
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
References
- 1.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. Candida auris sp.nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiology and immunology. 2009;53(1):41–4. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Chowdhary A, Sharma C, Duggal S, et al. New clonal strain of Candida auris, Delhi, India. Emerging infectious diseases. 2013;19(10):1670–3. doi: 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowdhary A, Anil Kumar V, Sharma C, et al. Multidrug-resistant endemic clonal strain of Candida auris in India. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2014;33(6):919–26. doi: 10.1007/s10096-013-2027-1. [DOI] [PubMed] [Google Scholar]
- 4.Lee WG, Shin JH, Uh Y, et al. First three reported cases of nosocomial fungemia caused by Candida auris. Journal of clinical microbiology. 2011;49(9):3139–42. doi: 10.1128/JCM.00319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emara M, Ahmad S, Khan Z, et al. Candida auris candidemia in Kuwait, 2014. Emerging infectious diseases. 2015;21(6):1091–2. doi: 10.3201/eid2106.150270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magobo RE, Corcoran C, Seetharam S, Govender NP. Candida auris-associated candidemia, South Africa. Emerging infectious diseases. 2014;20(7):1250–1. doi: 10.3201/eid2007.131765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvo B, Melo AS, Perozo-Mena A, et al. First report of Candida auris in America: Clinical and microbiological aspects of 18 episodes of candidemia. The Journal of infection. 2016;73:369–74. doi: 10.1016/j.jinf.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Morales-Lopez SE, Parra-Giraldo CM, Ceballos-Garzon A, et al. Invasive Infections with Multidrug-Resistant Yeast Candida auris, Colombia. Emerging infectious diseases. 2017;23(1):162–4. doi: 10.3201/eid2301.161497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lockhart SR, Etienne KA, Vallabhaneni S, et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2017;64(2):134–40. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vallabhaneni S, Kallen A, Tsay S, et al. Investigation of the First Seven Reported Cases of Candida auris, a Globally Emerging Invasive, Multidrug-Resistant Fungus - United States, May 2013–August 2016. MMWR Morbidity and mortality weekly report. 2016;65(44):1234–7. doi: 10.15585/mmwr.mm6544e1. [DOI] [PubMed] [Google Scholar]
- 11.Ruiz Gaitan AC, Moret A, Lopez Hontangas JL, et al. Nosocomial fungemia by Candida auris: First four reported cases in continental Europe. Revista iberoamericana de micologia. 2017 doi: 10.1016/j.riam.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Schelenz S, Hagen F, Rhodes JL, et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrobial resistance and infection control. 2016;5:35. doi: 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cendejas-Bueno E, Kolecka A, Alastruey-Izquierdo A, et al. Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeasts. Journal of clinical microbiology. 2012;50(11):3641–51. doi: 10.1128/JCM.02248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma C, Kumar N, Meis JF, Pandey R, Chowdhary A. Draft Genome Sequence of a Fluconazole-Resistant Candida auris Strain from a Candidemia Patient in India. Genome announcements. 2015;3(4) doi: 10.1128/genomeA.00722-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chatterjee S, Alampalli SV, Nageshan RK, Chettiar ST, Joshi S, Tatu US. Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genomics. 2015;16:686. doi: 10.1186/s12864-015-1863-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prakash A, Sharma C, Singh A, et al. Evidence of genotypic diversity among Candida auris isolates by multilocus sequence typing, matrix-assisted laser desorption ionization time-of-flight mass spectrometry and amplified fragment length polymorphism. Clin Microbiol Infect. 2016;22(3):277 e1–9. doi: 10.1016/j.cmi.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 17.Ben-Ami R, Berman J, Novikov A, et al. Multidrug-Resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerging infectious diseases. 2017;23(1) doi: 10.3201/eid2302.161486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizusawa M, Miller H, Green R, et al. Journal of Clinical Microbiology. JCM; 2016. Can a multi-drug resistant Candida auris be reliably identified in clinical microbiology laboratories? pp. 02202–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kathuria S, Singh PK, Sharma C, et al. Multidrug-Resistant Candida auris Misidentified as Candida haemulonii: Characterization by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry and DNA Sequencing and Its Antifungal Susceptibility Profile Variability by Vitek 2, CLSI Broth Microdilution, and Etest Method. Journal of clinical microbiology. 2015;53(6):1823–30. doi: 10.1128/JCM.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark AE, Kaleta EJ, Arora A, Wolk DM. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clinical microbiology reviews. 2013;26(3):547–603. doi: 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girard V, Mailler S, Chetry M, et al. Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses. 2016;59:535–8. doi: 10.1111/myc.12519. [DOI] [PubMed] [Google Scholar]
- 22.Bizzini A, Greub G. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2010;16(11):1614–9. doi: 10.1111/j.1469-0691.2010.03311.x. [DOI] [PubMed] [Google Scholar]
- 23.Dingle TC, Butler-Wu SM. Maldi-tof mass spectrometry for microorganism identification. Clinics in laboratory medicine. 2013;33(3):589–609. doi: 10.1016/j.cll.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Pfaller MA, Castanheira M, Lockhart SR, Ahlquist AM, Messer SA, Jones RN. Frequency of decreased susceptibility and resistance to echinocandins among fluconazole-resistant bloodstream isolates of Candida glabrata. Journal of clinical microbiology. 2012;50(4):1199–203. doi: 10.1128/JCM.06112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cleveland AA, Farley MM, Harrison LH, et al. Changes in incidence and antifungal drug resistance in candidemia: results from population-based laboratory surveillance in Atlanta and Baltimore, 2008–2011. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2012;55(10):1352–61. doi: 10.1093/cid/cis697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vincent BM, Lancaster AK, Scherz-Shouval R, Whitesell L, Lindquist S. Fitness trade-offs restrict the evolution of resistance to amphotericin B. PLoS Biol. 2013;11(10):e1001692. doi: 10.1371/journal.pbio.1001692. [DOI] [PMC free article] [PubMed] [Google Scholar]