Abstract
Opportunistic fungal infections are a leading cause of death for immune-compromised patients and there is pressing need to develop new anti-fungal therapeutic agents because of toxicity and resistance to current anti-fungal drugs. Although C-type lectin receptor- and Toll-like receptor-induced signaling pathways are key activators of host anti-fungal immunity, little is known about the negative regulation of these immune responses. Here, we found that JNK1 activation suppresses anti-fungal immunity in mice. We showed that JNK1-deficient mice had significantly higher survival rate in response to Candida albicans infection, and JNK1 expressed in hematopoietic innate immune cells is critical for this effect. JNK1 deficiency leads to significantly higher induction of CD23, a novel C-type lectin receptor, through NFATc1-mediated regulation of the CD23 promoter. Blocking CD23 upregulation or CD23-dependent nitric oxide production eliminated the enhanced anti-fungal effect in JNK1-deficient mice. Notably, JNK inhibitors exerted potent anti-fungal therapeutic effects in Candida albicans-infected mouse and human cells, indicating that JNK1 can be a therapeutic target for treating fungal infection.
Invasive fungal infection kills about one and a half million people every year worldwide1,2. Candida albicans is the most frequent fungal species isolated from infected patients3. Increasing numbers of immuno-compromised patients, including HIV-infected individuals, organ transplant recipients, and cancer patients treated with chemotherapy, limited numbers of anti-fungal drugs, and drug resistance are the main reasons for the high morbidity and mortality associated with disseminated candidiasis1,4. Therefore understanding how the host immune system fights fungal infections is crucial to develop novel immune response-based therapies2,5.
Pattern recognition receptors, including Toll-like receptor (TLR), C-type lectin receptor (CLR), Nod-like receptor (NLR) and RIG-I-like receptor (RLR), initiate the host immune response against invading pathogens 6. Previous studies indicate that CLRs play critical roles in recognizing fungal surface components leading to induction of host anti-fungal immune responses5–8. The CLRs Dectin-1, Dectin-2, Dectin-3 (also named MCL), recognize various carbohydrate, glycoprotein or glycolipid components of the fungal cell wall, such as β-glucan or α-mannan, which trigger the downstream signaling cascades essential for protective immunity against fungi 9–14. Activation of spleen tyrosine kinase (Syk) through CLRs triggers CARD9-BCL10-MALT1 (CBM) complex-dependent NF-κB signaling in macrophages or dendritic cells (DCs), and results in the release of pro-inflammatory cytokines, including tumor necrosis factor alpha (TNFα), interleukin (IL)-6, and IL-17, among others15,16. Phagocytosis, reactive oxygen species (ROS) production, neutrophil recruitment, and inflammasome activation have been shown to play critical roles in the fungal killing process6–8. Recently, three groups including us reported that the activated CLRs are rapidly targeted to lysosome-mediated degradation in response to fungal infection17–19.
c-Jun N-terminal kinases (JNKs) play important roles in T cell activation and T helper cell differentiation, cell apoptosis, obesity, insulin resistance, and tumorigenesis20–23. Many efforts have identified various ATP-competitive or ATP-noncompetitive JNK inhibitors24,25. Although a few studies have shown that JNK can be activated by various pattern recognition receptors22, the functional roles of JNK in innate immune responses have not been well characterized. In particular, the role of JNK activation in host anti-fungal responses has not been studied. Here we report that JNK1 negatively regulates the host anti-fungal innate immune response through suppressing CD23 expression, and may serve as a therapeutic target against fungal infection.
RESULTS
JNK1 negatively regulates the host anti-fungal innate immune responses in vivo
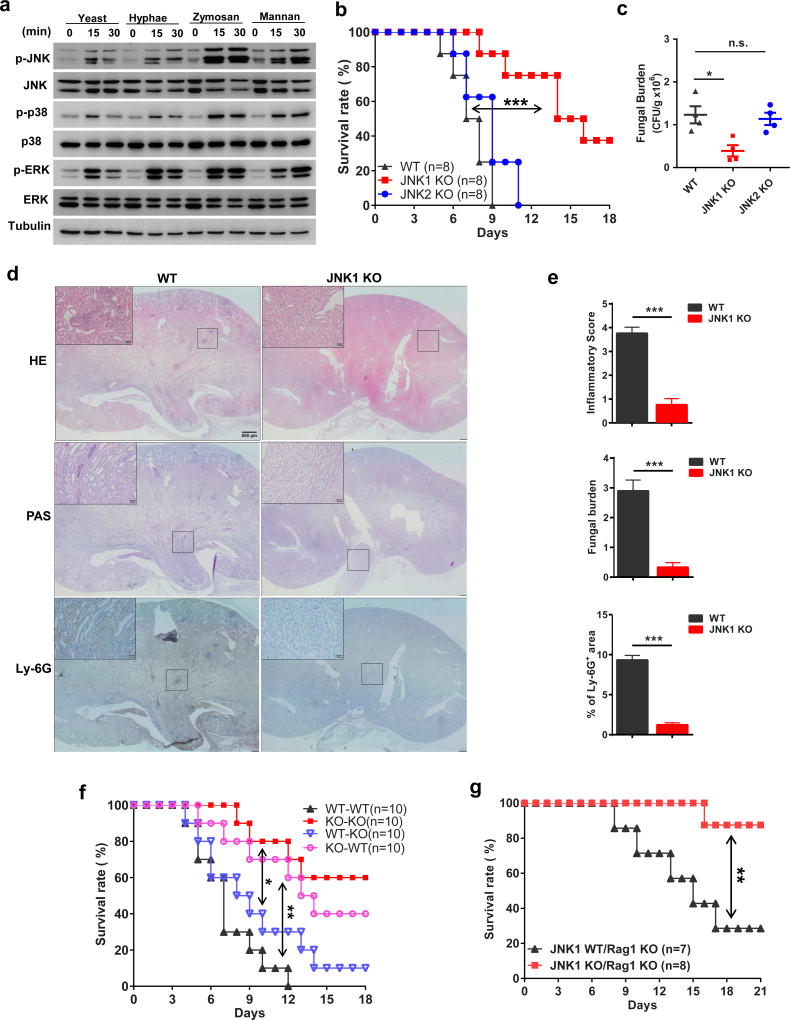
To investigate the role of JNK in fungal infection response, bone marrow-derived macrophages (BMDM) were stimulated with yeast or hyphae form of C. albicans, and the fungal cell wall component Zymosan or α-mannan. All these stimuli effectively induced JNK1/2 phosphorylation (Fig. 1a). To further characterize JNK function in the host immune response against fungal infection, we intravenously infected JNK1 knockout (KO), JNK2 KO, and wild type (WT) control mice with a sub-lethal dose of C. albicans. We found that JNK1 KO mice were more resistant to fungal infection compared with JNK2 KO or WT mice (Fig. 1b). Consistently, the fungal load in the kidneys was significantly lower (Fig. 1c and Supplementary Fig. 1a). JNK1 KO mice also manifested reduced renal inflammation and reduced numbers of C. albicans colony forming units (CFU) in the kidney (Fig. 1d, e and Supplementary Fig. 1b, c). The above findings were confirmed using two different doses of fungal infections in JNK1 KO and littermate heterozygous mice (Supplementary Fig. 1d, e). These data suggest that deficiency of JNK1 but not JNK2 in the host leads to a boost in antifungal immunity.
Figure 1. JNK1 negatively regulates the antifungal innate immune response.
(a) Wildtype BMDMs were stimulated with yeast (MOI=10) and hyphae (MOI=0.1) form of C. albicans, plate-coated zymosan (100ug/ml) and α-mannan (100ug/ml) for the indicated time. Cell lysates were analyzed by immunoblotting with the indicated antibodies. (b, c) JNK1 knockout (JNK1 KO, n=8), JNK2 knockout (JNK2 KO, n=8) and control wildtype (WT, n=8) mice were intravenously injected with 2×105 CFU of C. albicans per mouse. Mice survival was monitored and plotted as shown in (b). Kidney fungal loading was assayed at day 2 after infection. Each dot represents a single mouse, n=4 for each group (c). (d, e) Kidney sections of the C. albicans infected JNK1 KO and WT mice were stained with haematoxylin and eosin (H&E), periodic-acid-Schiff (PAS), or Ly-6G. Insets show regions of fungal inflammation, regions of fungal growth, and regions of neutrophil infiltration, respectively. Representative images (d), combined inflammatory score based on renal immune cell infiltration and tissue destruction, fungal load score, and neutrophil marker Ly-6G were shown (e). n=3 for each group and three sections per kidney were analyzed. Insets show higher-magnification images of boxed areas; scale bars, 500µm, 50µm (insets). (f) Bone-marrow cells from JNK1 KO (n=10) and WT mice (n=10) were intravenously injected into the irradiated recipient mice separately. Seven weeks later, mice were intravenously infected with 2×105 CFU of C. albicans. Survival of these mice was monitored. (g) JNK1WT/Rag1KO (n=7) and JNK1KO/Rag1KO (n=8) mice were intravenously infected with 2×105 CFU of C. albicans per mouse. Survival of these mice was monitored. Statistical significance was calculated by Log-rank (Mantel-Cox) test, Two-tailed unpaired t test (c), Multiple t test (e). Data are mean ± SEM (c, e). n.s. P > 0.05, *P < 0.05, ** P < 0.01, *** P < 0.001.
Myeloid lineage cells, including macrophage and DC, are key effector cells against fungal during the first few days after initial infection3,26. JNK1 has been reported to be expressed on all of different tissue compartments22. To investigate the cellular basis of the JNK1-related antifungal effect, we generated bone marrow (BM)-chimeric mice by reconstituting lethally irradiated WT mice with syngeneic JNK1 KO BM, or JNK1 KO mice with WT BM. Loss of JNK1 in hematopoietic cells showed the similar phenotype with total JNK1 deficiency in response to C. albicans infection (Fig. 1f and Supplementary Fig. 1f, KO-WT vs. KO-KO). Hematopoietic cells contain both innate and adaptive immune cells. To determine the contribution of JNK1 in the adaptive immune system to the observed phenotype, we generated JNK1/Rag1-double knockout (KO) mice by crossing JNK1 KO mice with Rag1 KO mice. Notably, JNK1 deficiency in the absence of adaptive immune cells closely resembled the phenotype of whole-body JNK1 KO mice in response to C. albicans infection (Fig. 1g). Together, these data indicate that JNK1 negatively regulates the anti-fungal innate immune response in vivo.
JNK1 deficiency induce higher CD23 expression and nitric oxide production in vitro
Pro-inflammatory cytokines, such as IL-6 and TNFα, are reported to be key factors for the host innate immune system to fight against fungi5,26. However, we did not detect significant changes in the levels of these pro-inflammatory cytokines post-infection in JNK1 KO mice compared with WT mice using Multiplex cytokine array analysis (Supplementary Fig. 2a), which we confirmed in vitro using BMDM stimulated with C. albicans (Supplementary Fig. 2b). Activation of p38 and ERK was also comparable in WT and JNK1 KO cells (Supplementary Fig. 2c). To identify gene(s) that may be responsible for resistance to fungal infection in JNK1 KO mice, we performed RNA-Seq analysis with WT and JNK1 KO BMDM after yeast form C. albicans stimulation (Supplementary Fig. 3a). Notably, we found a novel C-type lectin gene, Fcer2a, was significantly upregulated in stimulated JNK1 KO cells compared with WT control cells (Fig. 2a).
Figure 2. JNK1 suppresses CD23 induction by C. albicans.
(a) WT and JNK1 KO BMDM cells were stimulated with yeast form C. albicans (MOI=10) for 24 hours or medium as control. Total RNA was extracted and RNA-Seq analysis was performed. Shown are the heatmaps of the selected gene panels. (b) JNK1 KO, JNK2 KO and control WT BMDM cells were stimulated with C. albicans yeast (MOI=10) for 48 hours. Cells were washed and collected for FACS staining. Shown are the CD23 staining within the gated CD11b positive cells. Experiments were performed in triplicates. (c, d) NIH-3T3 cells stably expressing CD23, Dectin-1, Dectin-3, or mock control vector were assayed for AFM with α-mannan (c) (20 mg/ml) or Curdlan (d) (10 mg/ml) pre-treated probe. Each dot indicates the max force of one cell touch with the coated probe. Representative result of two independent experiments was shown. (e) WT, JNK1 and JNK2 KO BMDM cells were stimulated with heat-inactivated C. albicans (MOI=10) for indicated time. NO production in culture supernatants at indicated time point was measured by Nitrite Assay kit. (f) P30A or control peptide (10ug/ml) was added together with C. albicans and cells were treated for 48 hours. NO production in culture supernatants was measured by Nitrite Assay kit. For panels (e, f) experiments were performed in triplicate. Statistical significance was calculated by Multiple t test (c, d, e, f). Data are mean ± SEM (c, d, e, f). n.s. P > 0.05, *P < 0.05, ** P < 0.01, *** P < 0.001.
Fcer2a encodes the protein CD23 that was identified as the low affinity receptor for IgE 27, and is also a C-type lectin receptor. It is located on chromosome 19p131 and forms a gene cluster with Cd209a, which encodes DC-SIGN28. To validate the RNA-Seq data, we analyzed expression of a series of C-type lectin receptor genes by quantitative real time PCR, and found only Fcer2a was significantly induced in JNK1 KO BMDM cells (Supplementary Fig. 3b). We confirmed CD23 protein upregulation on the cell surface upon C. albicans stimulation by FACS (Fig. 2b). Since CD23 is also a C-type lectin receptor, we examined whether it can recognize surface components of fungi directly by performing a cellular binding assay. Indeed, we found that CD23 can bind both yeast and hyphae forms of C. albicans (Supplementary Fig. 4a). Since the C. albicans cell wall is mainly composed of α-mannan and β-glucan, we perform Atomic force microscopy (AFM) and ligand-binding assay to detect whether CD23-expressing cells or CD23 protein can bind with α-mannan and β-glucan directly. Indeed, we found that both cell surface expressed CD23 and purified soluble CD23 protein could effectively bind to α-mannan and β-glucan/Curdlan similarly to the known CLR receptors Dectin-1 and Dectin-3 (Fig. 2c, d and Supplementary Fig. 4b).
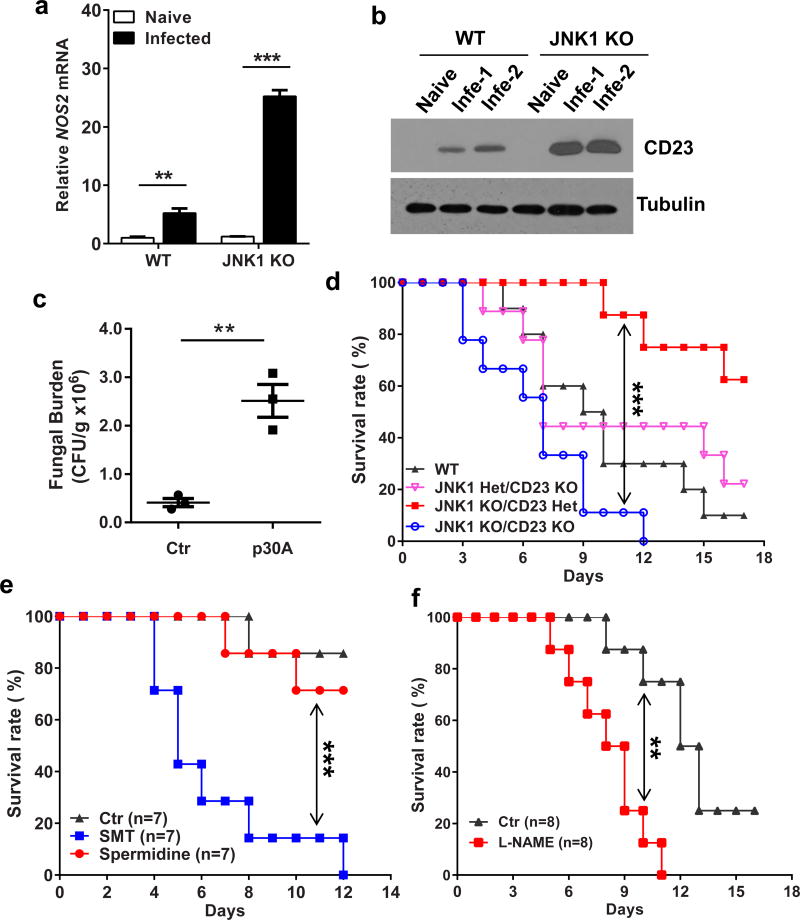
It has been reported that infection of human monocyte-derived macrophages with Mycobacterium avium induced membrane expression of CD23 29, and CD23 induces Nitric Oxide-Synthase (NOS) activity in monocytes 30,31. Furthermore, CD23 and Nitric Oxide (NO) is involved in the process killing Leishmania and Mycobacterium avium by human macrophages29,32,33. Notably, Nos2, which encodes the inducible NOS (iNOS), is highly differentially expressed between WT and JNK1 KO cells following C. albicans stimulation (Fig. 2a). Consistently, we found both α-mannan and β-glucan could stimulate CD23-overexpressing monocyte cell line, Raw264.7 cells, to produce more NO (Supplementary Fig. 4c). Importantly, JNK1 KO BMDM cells produced about three fold increased soluble NO in the supernatants compared with WT cells following C. albicans infection (Fig. 2e), which is consistent with the significant induction of Nos2 mRNA in JNK1 KO cells (Supplementary Fig. 5a). Furthermore, this elevated NO production in JNK1 KO cells is CD23 dependent, since JNK1 KO BMDM secreted much less NO upon treatment with p30A (Fig. 2f), a CD23-blocking peptide that effectively induce CD23 endocytosis from the cytoplasmic membrane34. p30A treatment decreased both the infected WT and JNK1 KO BMDM cell surface CD23 expression efficiently (Supplementary Fig. 5b), and treated JNK1 KO BMDMs could not kill C. albicans as efficiently as those treated with a control peptide (Supplementary Fig. 5c). Using of a NO synthesis inhibitor, L-NAME, to block NO production, also abolished the in vitro killing by JNK1 KO BMDM cells (Supplementary Fig. 5d, e). Together, these results show that JNK1 deficiency leads to the up-regulation of CD23 and higher NO production in fungal infected cells, which contribute to the enhanced fungal killing effect in the absence of JNK1.
Elevated CD23 and Nitric Oxide is essential for JNK1-related antifungal response in vivo
To confirm the function of CD23 and NO in vivo, we collected kidney tissues from C. albicans-infected mice, and found that Nos2 and Fcer2a mRNA expression was significantly higher in JNK1 KO mice (Fig. 3a and Supplementary Fig. 6a). CD23 protein level in the infected kidney was also higher in JNK1 KO mice (Fig. 3b), and p30A treatment could efficiently inhibit CD23 expression in mice (Supplementary Fig. 6b). To evaluate whether CD23 is responsible for the fungal protective effect in JNK1 KO mice, both blocking peptide and CD23 KO mice were used. Fungal burden of p30A-treated JNK1 KO mice was much higher than control peptide treated mice (Fig. 3c and Supplementary Fig. 6c), which is consistent with our in vitro data. JNK1/CD23-double knockout mice were generated by crossing CD23 KO mice with JNK1 KO mice, and they were infected with C. albicans. In accordance with the p30A blocking data, the higher survival rate in JNK1-single KO mice was impaired in JNK1/CD23-double KO mice (Fig. 3d and Supplementary Fig. 6d). Interestingly, the JNK1/CD23-double KO mice died even faster than WT and CD23-single KO mice for unknown reasons.
Figure 3. Elevated CD23 and iNOS expression in JNK1 KO mice are responsible for the enhanced antifungal immune response.
(a, b) WT and JNK1 KO mice (n=3 for each group) were intravenously infected with 5×105 CFU of C. albicans. 48 hours later, kidneys were isolated for total RNA. Nos2 was measured by quantitative real-time PCR (a). CD23 was detected by immunoblotting for kidney protein lysates (b). (c) JNK1 KO mice (n=3 for each group) were intravenously inoculated with 5×105 CFU of C. albicans per mouse. Infected mice were administered intraperitoneally with 200 mg/kg p30A or control peptide three times for 2 days. Mice kidneys were harvested. CFU assay was performed to detect the fungal loading. (d) WT (n=10), JNK1heterzygous (Het)/CD23 KO (n=9), JNK1 KO/CD23 Het (n=8), and JNK1 KO/CD23 KO mice (n=9) were infected with 2×105 C. albicans. Survival of these mice was monitored and plotted. (e, f) JNK1 KO mice were intravenously injected with 2×105 CFU of C. albicans per mouse. Infected mice (n=7 for each group) were administered intraperitoneally with 30mg/kg/day of SMT (NOS2 specific inhibitor) or 10mg/kg/day of spermidine (NOS1 specific inhibitor) or PBS (Ctr) daily (e). L-NAME (100 mg/kg) or PBS (Ctr) was administered intraperitoneally every day to the infected mice (n=8 for each group) (f). Survival of the mice was monitored and plotted. Statistical significance was calculated by Multiple t test (a), Two-tailed unpaired t test (c), and Log-rank (Mantel-Cox) test (d, f, e). Data are mean ± SEM (a, c). ** P < 0.01, *** P < 0.001.
Since iNOS expression is significantly elevated in response to fungal infection in JNK1 KO mice, we treated C. albicans-infected JNK1 KO mice with NOS1 specific inhibitor Spermidine and iNOS (NOS2) specific inhibitor SMT (Supplementary Fig. 6e). We found that SMT, but not Spermidine, significantly reduced survival rate of these mice (Fig. 3e). This iNOS specific effect was confirmed by using another inhibitor, L-NAME, which also reduced the survival rate of the infected JNK1 KO mice (Fig. 3f). These data indicate that CD23 induction and CD23-dependent elevation of nitric oxide are responsible for the enhanced anti-fungal immune response in JNK1 KO mice.
Monocytes/macrophages, dendritic cells, and neutrophils are all reported to be important for the anti-fungal immune response18,19,35. We examined which type of cells show increased expression of CD23 and iNOS in C. albicans-infected mice, and found that Fcer2a and Nos2 expression were significantly induced in bone marrow-derived macrophage and DC, but not in neutrophils in JNK1 KO mice after fungal infection (Supplementary Figure 7a). The neutrophil number decreased in JNK1 KO mice after fungal infection (Supplementary Figure 7b, c), which may be due to the lower fungal burden and reduced inflammatory response seven days after initial infection. Notably, peritoneal macrophages of CD23 KO mice express significantly less iNOS mRNA then control littermates (Supplementary Figure 7d). The fungal load was much higher in the kidneys of CD23 KO mice compared with WT mice after C. albicans infection (Supplementary Figure 7e). ROS production and phagocytosis are also important processes in anti-fungal responses19,36,37, and NO can interact with ROS and form peroxynitrite to efficiently kill fungi26,38. We found that JNK1-deficient cells produced higher amounts of ROS upon C. albicans infection (Supplementary Figure 8a), but the phagocytosis ability between wildtype and JNK1-deficient cells were comparable (Supplementary Figure 8b, c).
JNK1 negatively regulates CD23 expression through Dectin-1-induced NFAT activation
We next investigated how JNK1 deficiency results in upregulated CD23 expression following fungal infection. It has been shown that JNK1-deficient T cells exhibit elevated NFAT activation upon stimulation39, and NFAT bound to the CD23 promoter regulates its expression40,41. Interestingly, C. albicans stimulation can also trigger NFAT activation in macrophages and dendritic cells42,43. To determine the molecular mechanism by which CD23 expression is up-regulated following fungal infection, BMDM cells were stimulated with C. albicans and NFAT activation was examined. More NFATc1 translocated to the nucleus in JNK1 KO cells compared with WT cells upon C. albicans stimulation, although p65 nuclear translocation was comparable (Fig. 4a, b, c). Notably, JNK phosphorylation and NFAT activation was dependent on Dectin-1, since Dectin-1 KO BMDMs showed significantly reduced NFAT activation and JNK phosphorylation upon C. albicans yeast or Zymosan stimulation, whereas NF-κB subunit p65 translocation was not affected upon either α-mannan or Zymosan stimulation (Fig. 4d, e, and Supplementary Figure 9a, b, c, d). This result is consistent with the previous finding that Dectin-1 stimulation by C. albicans yeast or its ligand, Zymosan, triggers NFAT activation in macrophages42.
Figure 4. JNK1 negatively regulated CD23 expression through elevated Dectin-1 dependent NFAT activation.
(a–c) WT and JNK1 KO BMDM cells were stimulated with heat-inactivated C. albicans (MOI=10). Cells were collected after 1 hour stimulation and assay for immunofluorescence staining of NFAT-c1 (red) or p65 (green). Nuclei were counterstained with DAPI (blue). Magnification 200x, scale bars, 6.6µm (a). Statistical analysis of NFAT-c1 or p65 positive cells were shown, 12 images for each group were collected from three independent experiments for statistics (b). Nuclear extracts of the cells were collected at indicated time and analyzed by immunoblotting with NFAT-c1, NFAT-c2, p65 and PCNA antibodies (c). (d, e) WT, Dectin-1 KO and Dectin-3 KO BMDM cells were stimulated with α-mannan (10 ug/ml), Zymosan (20 ug/ml), or heat-inactivated C. albicans (MOI=10). Cells were collected after 1 hour stimulation and assay for immunofluorescence staining of NFAT-c1 or p65. Statistical analysis of NFAT-c1 (d) or p65 (e) positive cells was shown, 10 images for each group were collected from two independent experiments for statistics. (f) BMDMs from JNK1 KO mice were stimulated with heat-inactivated C. albicans (MOI=10) for 36 hours. TPCA-1 (NF-κB inhibitor, 1µM) or 11R-VIVIT (NFAT inhibitor, 10µM) was added to cells when stimulation began. After stimulation total RNA was extracted and CD23 expression was measured by quantitative real-time PCR, normalized to the internal control GAPDH. (g) ChIP assay with the indicated antibodies and real-time PCR analysis for CD23 promoter. Results are presented as means plus SEM after normalization to input. (h) HEK293T cells were co-transfected with the indicated CD23 promoter together with R-Luc. Twenty-four hours after transfection, cells were treated with PMA (200ng/ml)/Ionomycin (500ng/ml) for another 6 hours. NFAT inhibitor 11R-VIVIT (10uM) was added 20 minutes prior to PMA/Ionomycin stimulation. Luciferase assay were performed and R-Luc activity serves as internal control. For panels (f–h) experiments were performed in triplicate. Statistical significance was calculated by Multiple t test (b, d, e) or Two-tailed unpaired t test f (f, g, h). Data are mean ± SEM (b-h). n.s. P > 0.05, *P < 0.05, ** P < 0.01, *** P < 0.001.
To further determine whether CD23 induction is through a NFAT-dependent mechanism, we used 11R-VIVIT to inhibit NFAT activation, and then examined the expression level of CD23 in BMDM cells following fungal infection. We found that CD23 expression in JNK1 KO cells was significantly reduced upon 11R-VIVIT and C. albicans treatment (Fig. 4f). However, TPCA-1, which inhibits the NF-?B activation through IKK, hardly influences the CD23 induction. Since two putative NFAT-binding sites (-1368, and -714) were found in the Fcer2a promoter, we performed a ChIP assay and found a high level of binding of NFATc1 to the Fcer2a promoter in JNK1 KO cells following C. albicans stimulation (Fig. 4g). To determine whether the elevated nuclear translocation of NFATc1 dictates transcription of Fcer2a genes, we cloned the genomic promoter (about 1500bp) of Fcer2a and performed a luciferase reporter assay. We examined luciferase reporter activity, and found that the CD23 activation could be substantially induced when both potential NFAT binding sites were present, while adding the NFAT inhibitor, 11R-VIVIT, completely diminished this activation (Fig. 4h, -1500). Deletion of the -1368 NFAT site severely impaired CD23 promoter activation, but additional deletion of the -714 NFAT site did not further reduce the promoter activity (Fig. 4h, -750 and -480). These data indicate that NFATc1 regulates CD23 expression through directly binding to the -1368 binding site in CD23 promoter.
JNK inhibitors promote the anti-fungal immune response in vivo and in vitro
SP600125 is one of the most extensively used ATP-competitive JNK inhibitors and several studies using SP600125 have demonstrated its potential for therapeutic intervention by directly inhibiting JNK 24,25,44. To investigate whether JNK inhibitors can efficiently boost the anti-fungal innate immune response, wild-type mice were infected with C. albicans and then treated with two different doses of SP600125. Administration of both doses of SP600125 increased the survival rate compared with the control mice (Fig. 5a). Consistently, JNK inhibitor treatment also significantly reduced the fungal burden (Fig. 5b), and increased NOS2 and CD23 expression in the kidney of infected mice (Fig. 5c, d). Moreover, SP600125 specifically inhibited JNK activation and had no additional effect on phosphorylation of other MAPK kinases (p38 and ERK) (Supplementary Fig. 10a, b). To confirm the anti-fungal effect of the JNK inhibitor in vitro, BMDM cells were pretreated with SP600125 and then stimulated with C. albicans. Cell surface staining showed that significantly higher CD23 expression was induced upon JNK inhibition (Supplementary Fig. 10c). Consistently, SP600125-treated cells produced more nitric oxide (Supplementary Fig. 10d) and more efficiently killed fungi (Supplementary Fig. 10e).
Figure 5. JNK inhibitor shows therapeutic effect for antifungal immunity in vivo.
(a–d) Wildtype C57BL/6 mice (n=10 for each group) were intravenously injected with 4×105 C. albicans and treated with 10mg/kg/day (Low, for b to d) or 30mg/kg/day (High) SP600125. Survival of these mice was monitored and plotted (a). Kidney fungal loading was performed at day 2 after infection. (b). Kidneys were isolated 48 hours post infection and extracted for total RNA and cell lysates. CD23 (Fcer2a) and iNOS (Nos2) expression was measured by quantitative real-time PCR (c). CD23 protein expression was detected by immunoblotting with the cell lysates (d). For panels (b-d), n=3 mice for each group were used. (e) Wildtype C57BL/6 mice (n=10 for each group) were intravenously infected with 3×105 C. albicans and treated with 10mg/kg/day JNK-IN-8. Survival of these mice was monitored and plotted. (f) Wildtype C57BL/6 mice (n=4 for each group) were intravenously infected with 5×105 C. albicans and 24 hours later injected intraperitoneally with 10mg/kg JNK-IN-8 once. Kidney fungal loading was performed after another two days after JNK-IN-8 treatment. (g) Wildtype C57BL/6 mice (n=8 for each group) were intravenously infected with 3×105 C. albicans and 24 hours later injected intraperitoneally with 10mg/kg/day SP600125 once for four continuous days. Survival of these mice was monitored and plotted. Statistical significance was calculated by Log-rank (Mantel-Cox) test (a, e, g), Two-tailed unpaired t test (b, c) and Multiple t test (f). Data are mean ± SEM (b, c, f). ** P < 0.01, *** P < 0.001.
To further confirm the anti-fungal effect of JNK inhibitor in vivo, we use another JNK inhibitor, JNK-IN-8 45, which specially targets JNK activation (Supplementary Fig. 10a, b), to treat the clinical C. albicans strain sc5314-infected mice. Strikingly, JNK-IN-8 shows even better anti-fungal protective effect than SP600125 (Fig. 5e). These results demonstrate that JNK inhibitors can promote anti-fungal immune responses both in vivo and in vitro.
JNK1 is a potential therapeutic target for antifungal infection
To investigate whether JNK inhibitor has a therapeutic benefit in fungal infection, we infected wildtype mice with C. albicans by intravenous injection, and 24 hours later, injected JNK-IN-8 via the tail vein. We found that a significantly higher fungal burden in kidneys of solvent-treated mice than those of mice treated with the JNK inhibitor (Fig. 5f). To further confirm the therapeutic effect of the JNK inhibitor, we first intravenously infected wildtype mice with C. albicans, and 24 hours later, treated the mice intraperitoneally with SP600125 or control solvent daily for four continuous days, and monitored the mortality of these mice. We found that most of the control mice died within ten days of infection, but mice treated with JNK inhibitor were significantly more resistant to infection (Fig. 5g). These results indicate that JNK inhibitors may serve as potent therapeutic drugs for enhancing host innate immunity against fungal infection.
To test the relevance of our results in mice to humans, we infected the human monocytic cell line THP-1 and human PBMC-derived monocytes with C. albicans. Both THP-1 and human primary cells treated with JNK inhibitor showed significant upregulation of CD23 and iNOS, and killed fungi more efficiently in vitro (Fig. 6a, b). Thus, we conclude that JNK1 deficiency leads to the up-regulation of C-type lectin receptor CD23 through Dectin-1 dependent NFAT hyper-activation, and the elevated CD23 expression induces the cells to produce more nitric oxide by directly recognizing the fungal cell wall components, which will efficiently eliminate the fungal infection (Fig. 6c). Therefore, the JNK inhibitor enhances antifungal immunity and may serve as a potential drug to protect from lethal C. albicans sepsis in clinic.
Figure 6. JNK inhibitor promotes the antifungal response in human cells.
(a) THP-1 cells were stimulated with C. albicans yeast (MOI=10) and treated together with SP600125 (40uM) for 24 or 48 hours. Total RNA were extracted from the resulted cells. CD23 and NOS2 gene expression was measured by RT-PCR. (b,c) Human PBMC-derived monocytes were stimulated with C. albicans yeast (MOI=10) and treated together with SP600125 (40uM) for 24 or 48 hours. Total RNA were extracted from the resulted cells. CD23 and NOS2 gene expression was measured by RT-PCR (b). Cell supernatants were collected and in vitro killing assay was performed (c). For panels (a, b) experiments were performed in triplicate. Statistical significance was calculated by Multiple t test (a, b). Data are mean ± SEM (a, b). *P < 0.05, ** P < 0.01, *** P < 0.001. (d) Proposed model for the mechanism by which JNK1 regulate the anti-fungal immune response. After initial infection, C. albicans is sensed by C type lectin receptor (Dectin-1). In the absence of JNK1, it will induce elevated NFATc1 activation, which bind to CD23 promoter and induce higher CD23 expression. And these lead to much more Nitric Oxide production to kill C. albicans. However in wildtype situation, physiological level of CD23 is induced.
DISCUSSION
In this study, we have revealed that JNK1 functions as a negative regulator in innate immune responses against fungal infection. We found that JNK1-deficient mice are more resistant to fungal infection than wild type mice, and JNK1 deficiency in myeloid cells is vital for this process. CD23, a novel C-type lectin receptor, is significantly induced in the absence of JNK1 following fungal infection, and plays a critical role in anti-fungal innate immunity. The higher level of CD23 induction in JNK1 KO cells is dependent on elevated NFATc1 activation, which binds to the CD23 promoter and induces CD23 expression in macrophages. The up-regulated CD23 in turn induces a much higher level of nitric oxide production that kills C. albicans. Notably, JNK inhibitors show a potent anti-fungal effect both in vitro and in vivo through CD23 up-regulation and induction of nitric oxide. Together, these findings suggest that JNK1 negative regulate the innate immune response against C. albicans infection, and the inhibition of JNK1 activation can enhance the innate immunity against fungal infection.
JNK is involved in many physiological and pathological processes. The mechanism by which JNK regulates T cell immune responses and T helper cell differentiation has been well studied by using mice deficient for JNK1 or JNK220. The role of JNK in survival signaling, cell death, cancer development and diabetes is well established21,24,25,46. Despite its activation by a wide range of biological processes, including growth factors, cellular or oxidative stress, inflammatory cytokines, and pathogens, the functional role of JNK in innate immunity, especially anti-pathogen responses, has not been characterized. For the first time, to our knowledge, we reveal that JNK1 functions as a negative regulator of the host response to infection by the fungal pathogen C. albicans. Importantly, our findings suggest potential utility of JNK inhibitors as a novel anti-fungal therapeutic approach.
Monocytes, which include macrophages, dendritic cells, and neutrophils, have been reported to play important roles in anti-fungal immune responses. During fungal infection, macrophages have a marked impact on the inflammatory environment by expression of iNOS, pro-inflammatory cytokines, and chemokines47. DCs can kill fungi directly or present fungal components to organ-draining lymph nodes to prime CD4 T cells47. When encountering infectious particles, neutrophils initiate an anti-microbial killing program that includes phagocytosis and production of various toxic agents, including ROS48. However, our studies find that CD23-dependent iNOS expression in JNK1 KO mice after fungal infection is significantly elevated in macrophage and DC, but not in neutrophils. Interestingly, the number of neutrophils decreases in JNK1 KO mice upon infection, which may be due to the lower fungal burden and reduced inflammatory responses seven days after initial infection. These data highlight the importance of nitric oxide produced by macrophages in antifungal immunity. However, the role of CD23 in host defense and its regulation of iNOS expression remain to be further investigated.
There is increasing evidence indicating that C-type lectin receptors recognize the microbial surface components and induce host anti-microbial innate immune responses. Several CLRs have been recently characterized that sense both fungal and mycobacteria infections. For example, Dectin-1 recognize C. albicans yeast cells by binding to surface β-glucans, while Dectin-2 forms a heterodimer with Dectin-3 to sense the hyphae form through α-mannan. However, the function of other structure-based C-type lectin receptor family members needs to be characterized. In our study, we reported that CD23, a novel C-type lectin receptor, which is well known for its function as a low affinity receptor for IgE, plays an unexpected role in the anti-fungal innate immune responses through recognition of surface components of fungal pathogens. We have found that CD23 is an inducible CLR and can recognize fungal surface components, α-mannan and β-glucan, which regulate anti-fungal innate immunity. Consistent with the role of CD23 as a novel pattern recognition receptor involved in anti-fungal innate immune responses, we have found that modulation of CD23 by either peptide inhibitors or genetic deletion of the gene encoding CD23 has significant impacts on anti-fungal innate immunity. Our data also indicate that CD23 regulates NOS2 expression, which likely contributes to its anti-fungal innate immunity. Together, these results suggest that CD23 has other functions besides its well-known role as a low affinity receptor for IgE.
In summary, our findings shed new light on the negative regulation of immune response during fungal infection, and may have far-reaching and translationally relevant implications for developing novel approaches to fight against fungal infection.
ONLINE METHODS
Antibodies and reagents
Antibodies against phospho-p38 (4631), phospho-ERK (9101), phospho-JNK (9251), and total p38 (9212), total JNK (9252) were purchased from Cell Signaling Technology; antibodies against p65 (sc-8008), PCNA (proliferating cell nuclear antigen) (sc-56), NFAT-c1 (sc-7294), NFAT-c2 (sc-7296), CD23 (sc-271900), ERK (sc-154), I?Bα (sc-371), and tubulin (sc-8035) were from Santa Cruz Biotechnology. Dectin-1 and Dectin-3 monoclonal antibody was generated by using the extracellular domain of Dectin-1or Dectin-3 as immunogens, which was described before11. TPCA-1 (T1452) and α-mannan (M3640) were purchased from Sigma. Zymosan (#tlrl-zyn) was got from Invivogen. NFAT inhibitor 11R-VIVIT (#13855), JNK inhibitor SP600125 (#10010466), NO inhibitor L-NAME (#80210) were got from Cayman chemical. JNK inhibitor JNK-IN-8 (#s4901) was purchased from Selleck. NOS1 inhibitor Spermidine (#s0010), iNOS inhibitor S-Methylisothiourea hemisulfate salt (SMT, #s0008) were from Beyotime Biotechnology. PerCP-Cyanine5.5 anti-mouse CD11b antibody (#45-0112-82) was purchased from eBioscience, FITC anti-mouse F4/80 (#123108) and PE anti-mouse CD23 (#101608) antibodies were from Biolegend, APC-Cy7 anti-mouse CD11b was from BD Pharmingen. FITC-conjugated goat anti-mouse IgG (H+L) secondary antibody was purchased from Invitrogen (#62-6511). CD23 blocking peptide (p30A, FHENWPS) and control peptide (pCtr, SFNYNYA) were synthetized by Peptide 2.0 Inc. (Chantilly, VA, USA) and GL Biochem Ltd. (Shanghai, China).
Expression plasmids
Mouse CD23, Dectin-1 and Dectin-3 were amplified by PCR with cDNA of mouse BMDM as template. PCR amplifying fragment was inserted into a lentivirus vector, pRV3, between the ClaI and EcoRI sites. Extracellular domain of CD23, Dectin-1 and Dectin-3 were amplified by PCR with mouse BMDM as template. PCR amplifying fragments were inserted into an expression vector, pET28a, between the EcoRI and XhoI sites. The PCR primers used were listed in Supplementary Table 1.
Mice
JNK1 knockout mice, JNK2-knockout mice, and CD23-knockout mice were purchased from the Jackson Laboratory. To obtain JNK1−/−/CD23−/− or JNK1−/−/Rag−/− mice, JNK1−/− mice were paired with CD23−/− or Rag−/− mice, and subsequent intercross of their offspring led to the generation of the double knock-out mice. Wildtype C57BL/6 mice were purchased from Jackson Laboratory and breed in the facility. All mice were housed in the specific pathogen-free animal facility at MD Anderson Cancer Center and Tsinghua University. In all experiments described here, sex- and age-matched mice were used. All animal experiments were performed in compliance with institutional guidelines and according to the protocol approved by the Institutional Animal Care and Use Committee of The University of Texas MD Anderson Cancer Center and Tsinghua University.
Bone marrow-chimeric mice
Six-week-old recipient mice were lethally irradiated by X-ray (550 rad × 2), and then intravenously transferred with 5×106 bone-marrow leukocytes from indicated donors. Chimaeras were used for further experiments 7–8 weeks after the initial reconstitution.
Murine Systemic Candidiasis Model
For in vivo C. albicans infection, mice were injected via lateral tail veins with 200 ul of a suspension containing different doses of C. albicans (SC5314) in sterile PBS (Hyclone). Mouse survival rates were monitored following infection. Fungal loading was assessed by plating a series of diluted solutions of homogenized kidneys on the YPD plate.
Bone marrow-derived macrophage (BMDM) preparation
Primary cultures of BMDMs from indicated mice were prepared as previously described11. Briefly, bone marrow cells were harvested from the femurs and tibias of mice. Erythrocytes were removed from cells by using a hypotonic solution. Cells were cultured for 7 days in Dulbecco's modified Eagle medium containing 20% fetal bovine serum, 55 µM β-mercaptoethanol, streptomycin (100 µg/ml), penicillin (100 U/ml), and 30% conditioned medium from L929 cells expressing macrophage colony stimulating factor. After 6–7 days of culturing, flow cytometry analysis indicated that the harvested cell population contained above 97% CD11b+ F4/80+cells.
Isolation of neutrophils, monocytes, macrophages, and dendritic cells
Bone Marrow and splenic neutrophils, monocytes, macrophages, and dendritic cells were purified by CD45 MACS beads (Miltenyi, #130-052-301) first, and then FACS-sorting labeling with antibodies specific for CD11b (BD, #557396), CD11c (BD, #550261), Ly-6G (eBioscience, #48-5931-82), and Ly-6C (BD, #561237). Purified neutrophils, monocytes, macrophages, and dendritic cells were cultured in RPMI medium.
Immunoblotting assay
BMDMs were stimulated and lysed in lysis buffer (150 mM NaCl, 50 mM HEPES (pH 7.4), 1 mM EDTA, 1% Nonidet P-40, protease inhibitors). Nuclear extracts or total cell lysates were subjected to SDS-PAGE and then blotted using indicated antibodies.
Quantitative PCR
Total RNA was isolated using TRIZOL (Invitrogen) and reverse transcribed using SuperScriptIII (Invitrogen). Quantitative PCR was performed in triplicates using Power SYBR Green PCR Master Mix (Applied Biosystems). The amounts of transcript were normalized to GAPDH. Melting curves were run to ensure amplification of a single product. Primers used were listed in Supplementary Table 1.
RNA-seq
The RNA-sequencing was performed by Novogene (Beijing, China). Sequencing libraries were generated using NEBNext Ultra™ Directional RNA Library Prep Kit for Illumina; clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina); sequencing was done on an Illumina Hiseq platform and paired-end reads were generated. The filtered reads were aligned to the mouse most recent genome reference mm10 by using TopHat (version 2.1.0). After the alignment, the BAM files of each individual alignment were used to analyze genes differential expression by using Cufflinks (version 2.2.1). The heatmaps were generated by using gplots package in R (version 3.2.3). The RNA-Seq data is being deposited in NCBI Gene Expression Omnibus and the accession GEO number is GSE83265.
Chromatin immunoprecipitation assay
The Chromatin immunoprecipitation (ChIP) assay was performed with a ChIP kit (Active Motif, Cat#53009) as previously described before49. The antibodies used for ChIP are as follows: NF?B-p65 (sc-109), NFATc1 (sc-7294). The resulting DNA was analyzed by real-time PCR. The PCR primers used were listed in Supplementary Table 1.
Cytokine Measurement
Cytokine panel in the mouse serum were detected with Milliplex Map kit (Millipore, #MCYTOMAG-70K). All assays were performed according to the manufacturer’s protocols, and the MFIs were detected by the Luminex 200 system and were analyzed with Bio-Plex software (Bio-Rad). TNF-α, IL-6, IL-1β levels in the supernatant of the cells were measured with Ready-SET-GO enzyme-linked immunosorbent assay (ELISA) kits (eBioscience). All samples were measured in triplicate according to the manufacturer’s protocol.
Luciferase reporter assay
Luciferase assay were performed as described previously49. Briefly, 293T cells were transfected with 200ng of luciferase (firefly) reporter plasmid pGL3 containing the different length of CD23 promoter together with 4ng of EF1α promoter-dependent Renilla luciferase reporter. The transfected cells were cultured in DMEM containing 10% FBS for about 24 hours. These cells were treated with PMA (200ng/ml)/ Inomycine (500ng/ml) for another 6 hours. NFAT inhibitor 11R-VIVIT (10uM) was added 20 minutes prior to PMA/Inomycine stimulation. Resulted cells were harvested and lysed. Firefly and renilla luciferase activity was measured with a Dual-Luciferase Reporter system (Promega) and renilla luciferase was used to normalize transfection efficiency and luciferase activity.
Immunofluorescence staining
Indicated cells were fixed with 4% paraformaldehyde (PFA), permeabilized with 0.25% Triton X-100 and blocked with 10% goat serum. NFAT-c1 (Sant Cruz, sc-109) and p65 (Sant Cruz, sc-7294) were used as the first antibodies, Cy3-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch, 715-166-150) and Alexa Fluor 488-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch, 711-546-152) were used as the secondary antibodies. DAPI (Beyotime Biotech) was counterstained to label the cell nuclear. Fluorescence was detected using a Zeiss LSM780 confocal laser scanning microscope.
Histopathology
For histopathology analyses, kidneys were fixed in 4% Paraformaldehyde solution, processed according to standard procedures, embedded in paraffin, and sectioned. 5um thick sections were stained with haematoxylin and eosin (H&E), periodic-acid-Schiff (PAS), Gomori Methenamine Silver (GMS), or anti-Ly-6G (MDL, MD6477-020). Stained slides were scanned using a Microscope (OLYMPUS, IX73) and evaluated for severity of inflammation and intra-lesional fungal burden as described previously19. Renal inflammation was scored based upon H&E and PAS stainings (proportion of renal parenchyma and/or pelvis involved by tubulointerstitial nephritis and/or pyelonephritis) as not significant (score 0), less than 10% (score 1), 10–25% (score 2), 25–50% (score 3), or greater than 50% (score 4). The intra-lesional fungal burden was based upon PAS and GMS stainings as not significant (score 0), scant presence in less than 10% of inflammatory foci (score 1), mild-to-moderate presence in 10–25% of inflammatory foci (score 2), moderate-to-significant presence in 25–50% of inflammatory foci (score 3), or significant presence in more than 50% of inflammatory foci (score 4). ImageJ software was applied to evaluate the extent of infiltration by Ly-6G+ cells in affected kidneys.
Detection of reactive oxygen (ROS) production
The production of ROS was assayed as described with minor modification36. Briefly, 2 × 105 BMDM cells were washed with PBS twice and re-suspended in DMEM containing 10uM DCFH-DA. Cells were incubated at 37 °C for 30 min, after incubation cells were washed with DMEM five times to remove the nonspecific binding. Then cells infected with heated killed C. albicans at MOI = 10 for different time points, respectively. The relative amount of ROS generated was detected every 10 min by flow cytometry measuring the mean fluorescence intensity (MFI) in FL1 channel.
Phagocytosis of C. albicans and in vitro fungal killing assay
The phagocytosis assay was performed as described37 with minor modification. Briefly, 5 × 105 FACS-sorted cells were dispensed into 24-well plate and incubated at 37 °C for 2 hours to create a monolayer of phagocytes, followed by gently washing with culture medium to remove non-adherent cells. Resulting cells were incubated with 5×105 CFU C. albicans (MOI=1) at 37 °C for 15 minutes, supernatants were collected and monolayers were washed gently with DMEM to remove uningested microorganisms. The supernatant and well washings, containing the non-phagocytized C. albicans were combined and plated in serial dilutions on YPD agar plates incubated overnight at 30 °C. The percentage of phagocytized microorganisms was defined as (1- (number of uningested CFU / CFU at the start of incubation)) × 100. For phagocytosis of C. albicans by BMDM cells, GFP-labelled C. albicans yeast was used50. BMDM cells were stained with APC-labelled CD11b antibody (Biolegend, #101211), and then co-cultured with UV crosslinked GFP-labelled C. albicans (MOI=5) for 1 hour at 37 °C. Adherent fungal cells were quenched with trypan blue, and gated CD11b+ cells phagocytosis rate was determined by flow cytometry with GFP positive. In vitro fungal killing assay was performed as described before with minor modification14. Briefly, cells were allowed to interact for 30 min at 30 °C with live C. albicans (Ratio: 0.4 yeast per macrophage). Unbound particles were removed and cells were returned to the incubator for 4 hour to allow fungal killing. Control plates were kept at 4 °C to provide a measure of live fungi in the wells. After incubation, medium was removed and cells were lysed by incubation for 5 min at 25 °C with water at pH 11. Lysis buffer was neutralized with excess PBS and CFU was determined by plating on YPD agar incubated overnight at 30 °C.
Nitric Oxide Concentration Assay
Indicated cell culture supernatants were collected and Nitrite concentration was measured by a Nitric Oxide Assay Kit (Cat#S0021) from Beyotime Biotechnology (Shanghai, China) based on the Griess reaction. All samples were measured in triplicate according to the manufacturer’s protocol.
Ligand binding assay
Ligand binding assay was performed as previously described11. In brief, ELISA plates were coated with α-mannan or β-glucan (40 mg/ml) overnight and then added with 100 ul/well renatured protein of recombinant protein at indicated concentrations. Bound proteins were detected by their respective mouse monoclonal antibodies following by HRP-conjugated goat anti-mouse IgG secondary antibody (ESAYBIO, Beijing. #BE0102). Tetramethylbenzidine (TMB) substrate solution from eBioscience ELISA kit was used and the reaction was stopped by 2N H2SO4. OD450 was read on a SpectraMax Plus 384 Microplate Reader.
Cellular binding assay
The cellular binding assay was performed as described with minor modification14,51. Briefly, RAW264.7 cells stably expressing Flag-tagged CD23, Dectin-1 or Dectin-3 were co-cultured with GFP labelled yeast (MOI=5) or hyphae (MOI=0.1) form of C. albicans for 10 minutes, and uncombined yeast (for yeast form binding) or Raw cells (for hyphae form binding) were removed by four times washing with medium. Then cells were fixed with 4% paraformaldehyde (PFA), permeabilized with 0.25% Triton X-100 and blocked with 10% goat serum. Flag (Abmart, #M20008) were used as the first antibodies, Cy3-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch, 715-166-150) were used as the secondary antibodies. DAPI (Beyotime Biotech) was counterstained to label the cell nuclear. Fluorescence was detected using a Zeiss LSM780 confocal laser scanning microscope.
Atomic force microscopy (AFM)
The experiments were performed as described before52 using JPK Cellhesion unit (JPK, Berlin, Germany) with minor modification. (Set point: 0.5 nN, pulling length: 50um, delay mode: constant force, contact time: 5s). Probes (Nanoworld, #ARROW-TL-50) were pre-coated with α-mannan (20 mg/ml) or Curdlan (10 mg/ml) for overnight. NIH-3T3 cells stably expressing CD23, Dectin-1, Dectin-3, or control vector were assayed by the pre-treated probe. The max force of each touch was normalized and analyzed using JPK image processing software.
Isolation of human monocyte-derived monocytes (MDM)
Human MDMs were generated as previously described with minor modification18. In brief, peripheral blood mononuclear cells (PBMCs) from healthy donors were isolated from heparinized blood on Lymphoprep™ (Stemcell, #07851), a Ficoll-sodium based Density Gradient Medium. Monocytes were then separated by the Monocyte Isolation Kit II (Miltenyi Biotec, #130-091-153) and cultured in RPMI 1640 medium at 37 °C. The protocol was approved by the Ethics Committee of Tsinghua University.
Statistical analysis
All values in the paper are given as mean ± SEM, unless stated otherwise. All in vitro experiments were reproduced at least 3 independent times and all in vivo experiments were reproduced more than twice unless stated otherwise. Statistical significance was calculated by Two-tailed unpaired t test, Multiple t test, or Log-rank (Mantel-Cox) test using GraphPad Prism software. Statistical significance was set based on the P value. n.s. P > 0.05, *P < 0.05, ** P < 0.01, *** P < 0.001.
Supplementary Material
Acknowledgments
We thank Dr. Xiaoyu Hu, Dr. Yingli Shang, and Dr. Ling Ni for helpful discussion. We thank Xia Xu, Libing Mu, Dr. Shan Xie and Dr. Tie Xia for technical assistance. We thank Daniel C. Lin for editing English of this manuscript. This work was partially supported by grants from the National Natural Science Foundation of China (81502460 and 31670904 to X.Q.Z., 91542107 and 81630058 to X.L., 81571611 to X.M.J) and National Institutes of Health (AI116722 to X.L.).
Footnotes
AUTHOR CONTRIBUTIONS
X. Z. and Y. G. performed most of experiments; C. J. and T. L. performed some of the mouse experiments; Q. C. and S. Z. performed some of the in vitro experiments; B. Z. analyzed the RNA-Seq data and helped preparing figures. X. J., M. -C.H. and C.D. provided key reagents and insightful discussion; X.L. and X. Z. conceived the project and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Supplementary information includes 10 figures and 1 table.
References
- 1.Brown GD, et al. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Kim JY. Human fungal pathogens: Why should we learn? J Microbiol. 2016;54:145–148. doi: 10.1007/s12275-016-0647-8. [DOI] [PubMed] [Google Scholar]
- 3.Wuthrich M, Deepe GS, Jr, Klein B. Adaptive immunity to fungi. Annu Rev Immunol. 2012;30:115–148. doi: 10.1146/annurev-immunol-020711-074958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gow NA, van de Veerdonk FL, Brown AJ, Netea MG. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nature reviews. Microbiology. 2012;10:112–122. doi: 10.1038/nrmicro2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Underhill DM, Pearlman E. Immune Interactions with Pathogenic and Commensal Fungi: A Two-Way Street. Immunity. 2015;43:845–858. doi: 10.1016/j.immuni.2015.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plato A, Hardison SE, Brown GD. Pattern recognition receptors in antifungal immunity. Seminars in immunopathology. 2014 doi: 10.1007/s00281-014-0462-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardison SE, Brown GD. C-type lectin receptors orchestrate antifungal immunity. Nature immunology. 2012;13:817–822. doi: 10.1038/ni.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoving JC, Wilson GJ, Brown GD. Signalling C-type lectin receptors, microbial recognition and immunity. Cellular microbiology. 2014;16:185–194. doi: 10.1111/cmi.12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown GD, et al. Dectin-1 is a major beta-glucan receptor on macrophages. The Journal of experimental medicine. 2002;196:407–412. doi: 10.1084/jem.20020470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson MJ, et al. Dectin-2 is a Syk-coupled pattern recognition receptor crucial for Th17 responses to fungal infection. The Journal of experimental medicine. 2009;206:2037–2051. doi: 10.1084/jem.20082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu LL, et al. C-type lectin receptors Dectin-3 and Dectin-2 form a heterodimeric pattern-recognition receptor for host defense against fungal infection. Immunity. 2013;39:324–334. doi: 10.1016/j.immuni.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 12.Saijo S, et al. Dectin-1 is required for host defense against Pneumocystis carinii but not against Candida albicans. Nature immunology. 2007;8:39–46. doi: 10.1038/ni1425. [DOI] [PubMed] [Google Scholar]
- 13.Saijo S, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32:681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Taylor PR, et al. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nature immunology. 2007;8:31–38. doi: 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dambuza IM, Brown GD. C-type lectins in immunity: recent developments. Current opinion in immunology. 2015;32:21–27. doi: 10.1016/j.coi.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gross O, et al. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature. 2006;442:651–656. doi: 10.1038/nature04926. [DOI] [PubMed] [Google Scholar]
- 17.Zhu LL, et al. E3 ubiquitin ligase Cbl-b negatively regulates C-type lectin receptor-mediated antifungal innate immunity. The Journal of experimental medicine. 2016;213:1555–1570. doi: 10.1084/jem.20151932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao Y, et al. Targeting CBLB as a potential therapeutic approach for disseminated candidiasis. Nat Med. 2016;22:906–914. doi: 10.1038/nm.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wirnsberger G, et al. Inhibition of CBLB protects from lethal Candida albicans sepsis. Nat Med. 2016;22:915–923. doi: 10.1038/nm.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Annu Rev Immunol. 2002;20:55–72. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
- 21.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 22.Arthur JS, Ley SC. Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol. 2013;13:679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- 23.Han MS, et al. JNK expression by macrophages promotes obesity-induced insulin resistance and inflammation. Science. 2013;339:218–222. doi: 10.1126/science.1227568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bogoyevitch MA, Ngoei KR, Zhao TT, Yeap YY, Ng DC. c-Jun N-terminal kinase (JNK) signaling: recent advances and challenges. Biochim Biophys Acta. 2010;1804:463–475. doi: 10.1016/j.bbapap.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Davies C, Tournier C. Exploring the function of the JNK (c-Jun N-terminal kinase) signalling pathway in physiological and pathological processes to design novel therapeutic strategies. Biochem Soc Trans. 2012;40:85–89. doi: 10.1042/BST20110641. [DOI] [PubMed] [Google Scholar]
- 26.Brown GD. Innate antifungal immunity: the key role of phagocytes. Annu Rev Immunol. 2011;29:1–21. doi: 10.1146/annurev-immunol-030409-101229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiwara H, et al. The absence of IgE antibody-mediated augmentation of immune responses in CD23-deficient mice. Proc Natl Acad Sci U S A. 1994;91:6835–6839. doi: 10.1073/pnas.91.15.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soilleux EJ, Barten R, Trowsdale J. DC-SIGN; a related gene, DC-SIGNR; and CD23 form a cluster on 19p13. J Immunol. 2000;165:2937–2942. doi: 10.4049/jimmunol.165.6.2937. [DOI] [PubMed] [Google Scholar]
- 29.Mossalayi MD, et al. CD23 mediates antimycobacterial activity of human macrophages. Infect Immun. 2009;77:5537–5542. doi: 10.1128/IAI.01457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aubry JP, et al. The 25-kDa soluble CD23 activates type III constitutive nitric oxide-synthase activity via CD11b and CD11c expressed by human monocytes. J Immunol. 1997;159:614–622. [PubMed] [Google Scholar]
- 31.Lecoanet-Henchoz S, et al. CD23 regulates monocyte activation through a novel interaction with the adhesion molecules CD11b-CD18 and CD11c-CD18. Immunity. 1995;3:119–125. doi: 10.1016/1074-7613(95)90164-7. [DOI] [PubMed] [Google Scholar]
- 32.Vouldoukis I, et al. IgE mediates killing of intracellular Toxoplasma gondii by human macrophages through CD23-dependent, interleukin-10 sensitive pathway. PLoS One. 2011;6:e18289. doi: 10.1371/journal.pone.0018289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vouldoukis I, et al. The killing of Leishmania major by human macrophages is mediated by nitric oxide induced after ligation of the Fc epsilon RII/CD23 surface antigen. Proc Natl Acad Sci U S A. 1995;92:7804–7808. doi: 10.1073/pnas.92.17.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rambert J, et al. Molecular blocking of CD23 supports its role in the pathogenesis of arthritis. PLoS One. 2009;4:e4834. doi: 10.1371/journal.pone.0004834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wirnsberger G, et al. Jagunal homolog 1 is a critical regulator of neutrophil function in fungal host defense. Nat Genet. 2014;46:1028–1033. doi: 10.1038/ng.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Underhill DM, Rossnagle E, Lowell CA, Simmons RM. Dectin-1 activates Syk tyrosine kinase in a dynamic subset of macrophages for reactive oxygen production. Blood. 2005;106:2543–2550. doi: 10.1182/blood-2005-03-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vonk AG, Wieland CW, Netea MG, Kullberg BJ. Phagocytosis and intracellular killing of Candida albicans blastoconidia by neutrophils and macrophages: a comparison of different microbiological test systems. J Microbiol Methods. 2002;49:55–62. doi: 10.1016/s0167-7012(01)00348-7. [DOI] [PubMed] [Google Scholar]
- 38.Romero-Puertas MC, Sandalio LM. Nitric Oxide Level Is Self-Regulating and Also Regulates Its ROS Partners. Front Plant Sci. 2016;7:316. doi: 10.3389/fpls.2016.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong C, et al. Defective T cell differentiation in the absence of Jnk1. Science. 1998;282:2092–2095. doi: 10.1126/science.282.5396.2092. [DOI] [PubMed] [Google Scholar]
- 40.Debnath I, Roundy KM, Weis JJ, Weis JH. Defining in vivo transcription factor complexes of the murine CD21 and CD23 genes. J Immunol. 2007;178:7139–7150. doi: 10.4049/jimmunol.178.11.7139. [DOI] [PubMed] [Google Scholar]
- 41.Kneitz C, et al. The CD23b promoter is a target for NF-AT transcription factors in B-CLL cells. Biochim Biophys Acta. 2002;1588:41–47. doi: 10.1016/s0925-4439(02)00114-x. [DOI] [PubMed] [Google Scholar]
- 42.Goodridge HS, Simmons RM, Underhill DM. Dectin-1 stimulation by Candida albicans yeast or zymosan triggers NFAT activation in macrophages and dendritic cells. J Immunol. 2007;178:3107–3115. doi: 10.4049/jimmunol.178.5.3107. [DOI] [PubMed] [Google Scholar]
- 43.Zanoni I, Granucci F. Regulation and dysregulation of innate immunity by NFAT signaling downstream of pattern recognition receptors (PRRs) Eur J Immunol. 2012;42:1924–1931. doi: 10.1002/eji.201242580. [DOI] [PubMed] [Google Scholar]
- 44.Bennett BL, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc Natl Acad Sci U S A. 2001;98:13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lalaoui N, et al. Targeting p38 or MK2 Enhances the Anti-Leukemic Activity of Smac-Mimetics. Cancer Cell. 2016;29:145–158. doi: 10.1016/j.ccell.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 46.Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Cell Biol. 2007;19:142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 47.Ersland K, Wuthrich M, Klein BS. Dynamic interplay among monocyte-derived, dermal, and resident lymph node dendritic cells during the generation of vaccine immunity to fungi. Cell Host Microbe. 2010;7:474–487. doi: 10.1016/j.chom.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borregaard N. Neutrophils, from marrow to microbes. Immunity. 2010;33:657–670. doi: 10.1016/j.immuni.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 49.Zhao XQ, et al. C-type lectin receptor dectin-3 mediates trehalose 6,6'-dimycolate (TDM)-induced Mincle expression through CARD9/Bcl10/MALT1-dependent nuclear factor (NF)-kappaB activation. J Biol Chem. 2014;289:30052–30062. doi: 10.1074/jbc.M114.588574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jia XM, et al. CARD9 mediates Dectin-1-induced ERK activation by linking Ras-GRF1 to H-Ras for antifungal immunity. The Journal of experimental medicine. 2014;211:2307–2321. doi: 10.1084/jem.20132349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sato K, et al. Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor gamma chain to induce innate immune responses. J Biol Chem. 2006;281:38854–38866. doi: 10.1074/jbc.M606542200. [DOI] [PubMed] [Google Scholar]
- 52.Flach TL, et al. Alum interaction with dendritic cell membrane lipids is essential for its adjuvanticity. Nat Med. 2011;17:479–487. doi: 10.1038/nm.2306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.