Abstract
Whole-genome sequencing (WGS) of bacterial isolates has become standard practice in many laboratories. Applications for WGS analysis include phylogeography and molecular epidemiology, using single nucleotide polymorphisms (SNPs) as the unit of evolution. NASP was developed as a reproducible method that scales well with the hundreds to thousands of WGS data typically used in comparative genomics applications. In this study, we demonstrate how NASP compares with other tools in the analysis of two real bacterial genomics datasets and one simulated dataset. Our results demonstrate that NASP produces similar, and often better, results in comparison with other pipelines, but is much more flexible in terms of data input types, job management systems, diversity of supported tools and output formats. We also demonstrate differences in results based on the choice of the reference genome and choice of inferring phylogenies from concatenated SNPs or alignments including monomorphic positions. NASP represents a source-available, version-controlled, unit-tested method and can be obtained from tgennorth.github.io/NASP.
Keywords: SNPs, Phylogeography, bioinformatics
Data Summary
No data was generated as part of this study.
Impact Statement
NASP represents a comprehensive, open-source method for SNP identification and differentiation between and among large numbers of microbial genomes. This method differs from other published SNP pipelines in terms of: (1) the variety of supported short-read aligners and SNP callers; (2) the variety of supported job management systems; (3) the ability to call both monomorphic and polymorphic sites; and (4) the ability to integrate the results from multiple SNP callers and identify the consensus set of SNPs that define the population structure. Accurate and comprehensive analysis of SNPs in a reference population is critical in outbreak investigations, source attribution and population genetics. NASP was developed for bacterial pathogens, but has also been used to analyze the population structure of fungal and viral pathogens. The NASP output can be used for genome-wide association studies (GWAS) to correlate the genotype and phenotype, and can also be used for phylogenomics, which allows for an understanding of the relatedness of microbial isolates across temporal and spatial scales.
Introduction
Whole-genome sequence (WGS) data from microbes, including bacteria, viruses, fungi and parasites, are rapidly increasing in public databases and have been used for outbreak investigations (Rasko et al., 2011; Eppinger et al., 2011; Engelthaler et al., 2016), associating phylogeny with serology (Sahl et al., 2015b) and phylogeography (Keim & Wagner, 2009; Engelthaler et al., 2014). WGS data are frequently used for variant identification, especially with regards to single nucleotide polymorphisms (SNPs). SNPs provide stable markers of evolutionary change between genomes (Foster et al., 2009). Accurate and reliable SNP identification requires the implementation of methods to call, filter and merge SNPs with tools that are version controlled, unit tested and validated (Olson et al., 2015).
Multiple pipelines are currently available for the identification of SNPs from diverse WGS datasets, although the types of supported input files differ substantially. There are few pipelines that support the analysis of both raw sequence reads as well as genome assemblies. The In Silico Genotyper (ISG) pipeline (Sahl et al., 2015a) calls SNPs from both raw reads, primarily from the Illumina platform, and genome assemblies, but isn’t optimized for job management systems and only exports polymorphic positions. While only polymorphic positions may be adequate for many studies, the inclusion of monomorphic positions in the alignment is important for calculating evolutionary rates. A commonly used SNP analysis software method is kSNP, which has been discussed in three separate publications (Gardner & Hall, 2013; Gardner & Slezak, 2010; Gardner et al., 2015). kSNP is a reference-independent approach in which all kmers of a defined length are compared to identify SNPs. The all-versus-all nature of the algorithm can result in a large RAM footprint and can stall on hundreds of bacterial genomes on some computational networks (Sahl et al., 2015a). Finally, REALPHY was published as a method to identify SNPs using multiple references and then merging the results (Bertels et al., 2014). The authors claim that single-reference-based methods bias the results, especially from mapping raw reads against a divergent reference genome.
Additional methods have also been published that only support specific input formats. Parsnp is a method that can rapidly identify SNPs from the core genome, but currently only processes closely related genome assemblies (Treangen et al., 2014). SPANDx is a method that only supports raw reads, but does run on a variety of job management systems (Sarovich & Price, 2014). The program lyve-SET has been applied to outbreak investigations and uses raw or simulated reads to identify SNPs (Katz et al., 2013). Finally, the CFSAN SNP pipeline is a published method from the United States Food and Drug Administration that only supports the use of raw reads (Pettengill et al., 2014). There have been, to our knowledge, no published comparative studies to compare the functionality of these pipelines on a range of test datasets.
In this study, we describe the NASP pipeline. NASP is a source-available, unit-tested, version-controlled method to rapidly identify SNPs and works on a range of job management systems, incorporates multiple read aligners and SNP callers, works on both raw reads and genome assemblies, calls both monomorphic and polymorphic positions, and has been validated on a range of diverse datasets. In this study, we compare NASP with other methods, both reference-dependent and reference-independent, in the analysis of three bacterial datasets.
Methods
NASP is implemented in a mixture of Python and Go programming languages. NASP accepts multiple file formats as input, including ‘.fasta’, ‘.sam’, ‘.bam’, ‘.vcf’, ‘.fastq’ and ‘fastq.gz’. NASP can either function through a question/answer command line interface designed for ease of use, or with a configuration file. NASP was developed to work on job management systems including Torque, Slurm and Sun/Oracle Grid Engine (SGE); a single-node solution is available for NASP as well, but is not optimal.
If filtering of duplicate regions in the reference genome is requested, the reference is aligned against itself with NUCmer (Delcher et al., 2003). These duplicated regions are then masked from downstream analyses, although still available for investigation. If external genome assemblies are supplied, they are also aligned against the reference genome with NUCmer and SNPs are identified by a direct one-to-one mapping of the query to the reference. In the case of duplications in the query but not the reference, all copies are aligned and any differences at any given base are masked with an ‘N’ character to identify it as ambiguous.
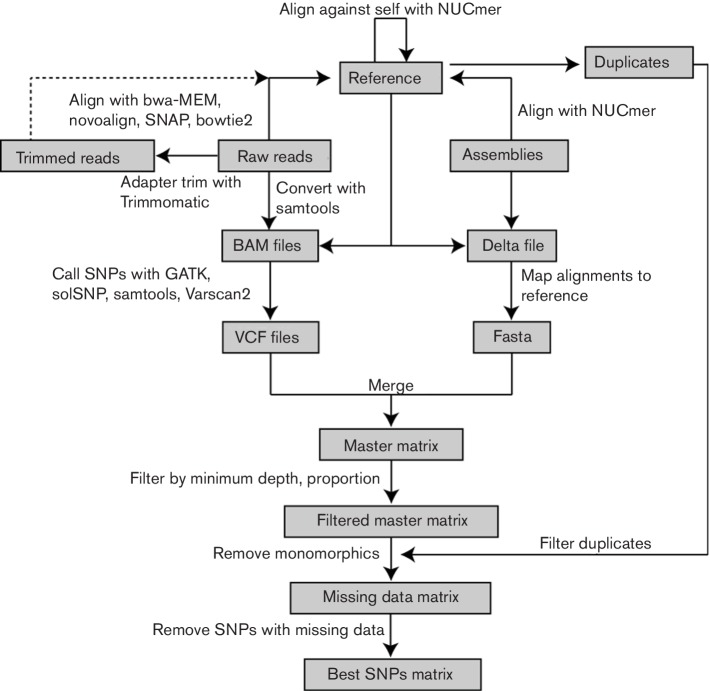
If raw reads are supplied, they can be adapter and/or quality trimmed with Trimmomatic (Bolger et al., 2014). Raw or trimmed reads are aligned against a FASTA-formatted reference using one or a combination of the supported short-read aligners, including BWA-MEM (Li, 2013), Novoalign (www.novocraft.com), bowtie2 (Langmead & Salzberg, 2012) and SNAP (Zaharia et al., 2011). A binary alignment map (BAM) file is created with Samtools (Li et al., 2009) and SNPs can be identified with multiple SNP callers, including the UnifiedGenotyper method in GATK (DePristo et al., 2011; McKenna et al., 2010), SAMtools, SolSNP (http://sourceforge.net/projects/solsnp/), and VarScan (Koboldt et al., 2012). If multiple aligners and/or SNP callers are selected, calls that are the same between all methods are reported in the bestsnp matrix as the consensus. Positions that fail a user-defined depth and proportion threshold (mixture of alleles) are filtered from downstream analyses but are retained in the ‘master’ matrices. A workflow of the NASP pipeline is shown in Fig. 1 and a summary is detailed in Table S1 (available in the online Supplementry Material).
Fig. 1.
Workflow of the NASP pipeline.
The results of the pipeline can include up to four separate SNP matrices. The first matrix is the master matrix (master.tsv), which includes all calls, both monomorphic and polymorphic, across all positions in the reference with no positions filtered or masked; positions that fall within duplicated regions are shown in this matrix, although they are flagged as duplicated. An optional second matrix (master_masked.tsv) can also be produced. This matrix is the same as the master matrix, although any position that fails a given filter (minimum depth, minimum proportion) is masked with an ‘N’, whereas calls that could not be made are given an ‘X’; this matrix could be useful for applications where all high-quality, unambiguous positions should be considered. The third matrix (missingdata.tsv) includes only positions that are polymorphic across the sample set, but can include those that are missing in a subset of genomes and not found in duplicated regions; these SNPs have also been processed with the minimum depth and proportion filters and are still high-quality calls. The last matrix (bestsnp.tsv) contains only polymorphic, non-duplicated, clean calls (A, T, C, G) that pass all filters across all genomes. FASTA files and multi-sample VCF files are automatically produced that correspond to the bestsnp and missingdata matrices.
In addition to the matrices, VCFs, and FASTA files, NASP produces statistics that can be useful for the identification of potentially problematic genomes, such as low-coverage or mixtures of multiple strains. These statistics can also be used for determining the size of the core, non-duplicated genome, including both monomorphic and polymorphic positions, of a given set of genomes.
Post matrix scripts are included with NASP in order to convert between file formats, remove genomes and/or SNPs, provide functional SNP information, and to convert into formats that can be directly accepted by other tools, such as Plink (Renteria et al., 2013), a method to conduct genome-wide association studies (GWAS). Documentation for all scripts is included in the software repository.
Test datasets.
To demonstrate the speed and functionality of the NASP pipeline, and to compare the output with other pipelines, three datasets were selected. The first includes a set of 21 genome assemblies of members of the genera Escherichia and Shigella used in other comparative studies (Bertels et al., 2014; Touchon et al., 2009) (Table S2). REALPHY was run on self-generated single-ended simulated reads, 100 bp in length. Additional pipelines were run with paired-end reads generated by ART chocolate cherry cake (Huang et al., 2012), using the following parameters: -l 100 -f 20 -p -ss HS25 -m 300 -s 50; this method was not run in conjunction with REALPHY, as the short-read generation is integrated into the method. Unless otherwise noted, the reference genome for SNP comparisons was Escherichia coli K-12 MG1655 (NC_000913) (Blattner et al., 1997). All computations were performed on a single node, 16-core server with 48 Gb of available RAM. For kSNP, the optimum k value was selected by the KChooser script included with the repository.
The second dataset includes a set of 15 Yersinia pestis genomes from North America (Table S3). For those external SNP pipelines that only support raw reads, simulated reads were generated from genome assemblies with ART. A set of SNPs (Table S4) has previously been characterized on these genomes with wet-bench methods (unpublished). This set was chosen to determine how many verified SNPs could be identified by different SNP pipelines. All computations were performed on a single node, 16-core server with 48Gb of available RAM.
The last dataset includes simulated data from Y. pestis. Reads and assemblies from 133 Y. pestis genomes (Cui et al., 2013) were downloaded from public databases and processed with NASP using the Colorado 92 (CO92) genome as the reference to produce a reference phylogeny for WGS data simulation. Assemblies and reads were simulated from this reference phylogeny and a reference genome (CO92 chromosome) using TreeToReads (https://github.com/snacktavish/TreeToReads), introducing 3501 mutations; in this process, mutations are introduced into genomes to reproduce the phylogeny, although the mutations are completely manufactured. A phylogeny was inferred from the concatenated SNP alignment (3501 simulated SNPs produced by TreeToReads) with RAxML v8 (Stamatakis, 2014) to provide a ‘true’ phylogeny for the simulated data. Simulated reads (250 bp) and assemblies were both processed with pipelines to identify how many of these introduced SNPs could be identified.
To test the scalability of NASP on genome assemblies, a set of 3520 E. coli genomes was selected (Table S5). Genomes were randomly selected with a python script (https://gist.github.com/jasonsahl/990d2c56c23bb5c2909d) at various levels (100–1000) and processed with NASP. In this case, NASP was run on multiple nodes across a 31-node high performance computing (HPC) cluster at Northern Arizona University. The elapsed time was reported only for the step where aligned files are compiled into the resulting matrix. Time required for the other processes is dependent on the input file type and the amount of available resources on a HPC cluster.
External SNP pipelines.
Multiple SNP pipelines, both reference-dependent and reference-independent, were compared with NASP, including kSNP v3.9.1 (Gardner et al., 2015), ISG v0.16.10–3 (Sahl et al., 2015a), Parsnp v1.2 (Treangen et al., 2014), REALPHY v112 (Bertels et al., 2014), SPANDx v3.1 (Sarovich & Price, 2014), Mugsy v1r2.2 (Angiuoli & Salzberg, 2011), lyve-SET v1.1.6 (Katz et al., 2013), and the CFSAN SNP pipeline (https://github.com/CFSAN-Biostatistics/snp-pipeline). Exact commands used to run each method are shown in Supplemental Data File 1. An overview of all tested methods is shown in Table 1. Most of the methods output FASTA or nexus files, which were used to infer phylogenies. For Mugsy, the MAF file was converted to FASTA with methods described previously (Sahl et al., 2011).
Table 1. An overview of commonly used SNP pipelines.
| Pipeline name | Supported data types | Output type | Parallel job management support? |
|---|---|---|---|
| NASP | FASTA, BAM, SAM, VCF, FASTQ, FASTQ.GZ | Matrix, VCF, FASTA | SGE, SLURM, TORQUE |
| ISG | FASTA, BAM, VCF, FASTQ, FASTQ.GZ | Matrix, FASTA | No |
| Parsnp | FASTA | gingr file, phylogeny, FASTA, VCF | No |
| REALPHY | FASTA*, FASTQ, FASTQ.GZ | FASTA, phylogeny | No |
| SPANDx | FASTQ.GZ | Nexus file, phylogeny | SGE, SLURM, TORQUE |
| CFSAN | FASTQ, FASTQ.GZ | SNP list, FASTA | SGE, TORQUE |
| kSNPv3 | FASTA | Matrix, FASTA, phylogeny | No |
| Mugsy | FASTA | MAF file | No |
| lyve-set | FASTQ.GZ, FASTA* | Matrix, FASTA, phylogeny | SGE |
*Generates simulated reads.
Phylogenetics.
Phylogenies were inferred using a maximum likelihood algorithm implemented in RAxML v8.1.7 (Stamatakis, 2014), except where noted. The exact commands used to infer the phylogenies are shown in Supplemental Data File 1. Tree topologies were also compared on the same input data using compare2trees (Nye et al., 2006). Commands to infer these phylogenies using FastTree2 v2.1.7 SSE3 (Price et al., 2010), ExaBayes v1.4.1 (Aberer et al., 2014), and Parsimonator v1.0.2 (github.com/stamatak/Parsimonator-1.0.2) are shown in Supplemental Data File 1.
Dendrogram of multiple methods.
To visually represent the performance of different methods, a dendrogram was generated. Each phylogeny was compared against a maximum likelihood phylogeny inferred from the reference test set with compare2trees and a congruence score was calculated. A unweighted pair group method with arithmetic mean (UPGMA) dendrogram was then calculated with Phylip v3.6 (Felsenstein, 2005) on the resulting similarity matrix.
RESULTS
Pipeline functionality and post-matrix scripts
NASP is a reference-dependent pipeline that can incorporate both raw reads and assemblies in the SNP discovery process; NASP was not developed for the identification and annotation of short insertions/deletions (indels). NASP can use multiple aligners and SNP callers to identify SNPs and the consensus calls can be calculated across all methods. A complete workflow of the NASP method is shown in Fig. 1. Several post-matrix scripts are included with NASP in order to convert between file formats, including generating input files for downstream pipelines [e.g. Plink (Renteria et al., 2013)]. An additional script can annotate a NASP SNP matrix using SnpEff (Cingolani et al., 2012) to provide functional information for each SNP.
NASP run time scalability
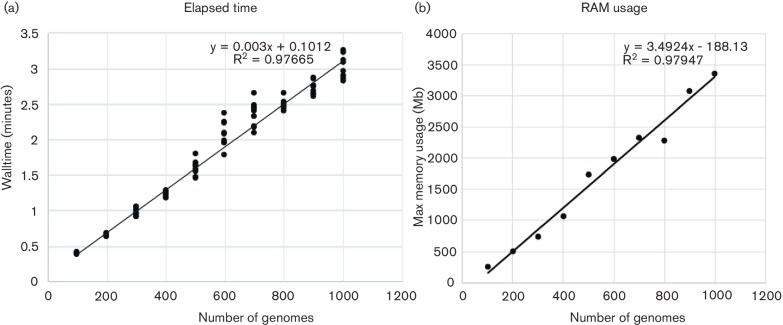
To visualize how NASP scales on processing genome assemblies, a set of 3520 E. coli genomes was sampled at 100-genome intervals and processed with NASP with 10 replicates. The results demonstrate that the matrix building step in NASP scales linearly up to three and a half minutes with the processing of additional genomes (Fig. 2a). The memory footprint of this step also scales linearly (Fig. 2b) and doesn’t exceed 4Gb on a large set of genomes (n = 1000). If raw reads are used, additional time is required for the alignment and SNP calling methods, and the overall wall time would scale with the number of reads that needed to be processed.
Fig. 2.
NASP benchmark comparisons of walltime (a) and RAM (b) on a set of Escherichia coli genomes. For the walltime comparisons, 3520 E. coli genomes were randomly sampled ten times at different depths and run on a server with 856 cores. Only the matrix-building step is shown, but demonstrates a linear scaling with the processing of additional genomes.
Pipeline comparisons on E. coli genomes data set
To test differences between multiple pipelines, a set of 21 genomes of members of the genera Escherichia and Shigella used in other comparative genomics studies (Bertels et al., 2014; Touchon et al., 2009) were downloaded and processed with Parsnp, SPANDx, kSNPv3, ISG, REALPHY, CFSAN, lyve-SET, Mugsy, and NASP. For methods that do not support genome assemblies, paired-end reads were simulated with ART, while single-end reads were used by REALPHY, as this method is integrated into the pipeline.
To identify how well the simulated paired end reads represent the finished genomes, a NASP run was conducted on a combination of completed genome assemblies as well as simulated raw reads. The phylogeny demonstrates that assemblies and raw reads fall into identical locations (Fig. S1, available in the online Supplementry Material), suggesting that the paired-end reads are representative of the finished genome assemblies.
The developers of REALPHY assert that their analysis of this dataset demonstrates the utility of using their approach to avoid biases in the use of a single reference genome by using multiple references (Bertels et al., 2014). To test differences between methods, SNPs were identified with multiple reference-dependent and -independent methods, and maximum likelihood (ML) phylogenies were compared with compare2trees. The results demonstrate that all methods, with the exception of kSNPv3 and lyve-SET, returned a phylogeny with the same topology as the published phylogeny (Bertels et al., 2014) (compare2trees topological score =100 %) (Table 2). The run wall time demonstrates that most other methods were significantly faster than REALPHY (Table 2), even when REALPHY was invoked using a single reference. Wall time comparisons between methods are somewhat problematic, as some pipelines infer phylogenies and others, including NASP, do not. Additionally, using raw reads is generally expected to be slower than using a draft or finished genome assembly. Finally, some methods are optimized for job management systems, whereas others were designed to run on a single node.
Table 2. SNP calling results on a set of 21 genomes of members of the genus Escherichia.
| Method | Reference | Data type | Parameters | Number of SNPs
considered |
Total number of sites | Walltime (single node - eight cores) | Topological score | Number of
defining SNPs |
|---|---|---|---|---|---|---|---|---|
| NASP | K12 MG1655 | Assemblies | Default | 267978* | 2322434 | 10 m 00 s | 100 % | 809 |
| NASP | K12 MG1655 | Assemblies | NUCmer (−b 20) | 162758* | 1839583 | 10 m 00 s | 100 % | 744 |
| NASP | K12 MG1655 | ART PE reads | BWA, GATK,
MinDepth = 3, MinAF = 0.90 |
170208* | 1984510 | 1 h 43 m 00 s | 100 % | 826 |
| NASP | E. fergusonii 35469 | Assemblies | Default | 244262* | 2227038 | 10 m 00 s | 100 % | 741 |
| NASP | E. fergusonii 35469 | ART PE reads | BWA, GATK,
MinDepth = 3, MinAF = 0.90 |
141238* | 1813349 | 1 h 17 m 10 s | 100 % | 748 |
| ISG | K12 MG1655 | Assemblies | Default | 268524* | N/A | 6 m 47 s | 100 % | 810 |
| ISG | K12 MG1655 | ART PE reads | minaf 0.9, mindp 3 | 206193* | N/A | 14 m 45 s | 100 % | 824 |
| Parsnp | K12 MG1655 | Assemblies | "-c d" | 151256* | 1682404 | 4 m 35 s | 100 % | 777 |
| REALPHY | K12 MG1655 | REALPHY SE reads | Default | 171828* | 1897146 | 3 h 11 m 00 s | 100 % | 779 |
| kSNPv3 | N/A | Assemblies | -core | 20587* | N/A | 27 m 58 s | 91.80 % | 5 |
| kSNPv3 | N/A | Assemblies | Default | 284134 | N/A | 27 m 58 s | 95.80 % | 547 |
| SPANDx | K12 MG1655 | ART PE reads | -t Illumina -p PE -z yes | 98492 | N/A | 3h 6 m | 100 % | 609 |
| CFSAN | K12 MG1655 | ART PE reads | Default | 128512* | N/A | 1 h 56 m 00 s | 100 % | 808 |
| Mugsy | N/A | Assemblies | Default | 307072* | 2478794 | 1 h 39 m 03 s | 100 % | unknown |
| lyve-SET | K12 MG1655 | ART PE reads | min_coverate 3, min_alt_frac 0.9 | 163118* | 1183153 | 6 h 25 m | 85 % | 329 |
*strictly core genome SNPs.
One of the other assertions of the REALPHY developers is that phylogenies reconstructed using an alignment of concatenated SNPs are unreliable (Bertels et al., 2014; Touchon et al., 2009), especially with regards to branch length biases (Leache et al., 2015). However, the phylogeny inferred from a NASP alignment of monomorphic and polymorphic sites was in complete agreement with the topology of the phylogeny inferred from a concatenation of SNPs (compare2trees topological score =100 %); tree lengths were indeed variable with use of these two different input types using the same substitution model (Fig. S2). We also employed an ascertainment bias correction (Lewis correction) (Leache et al., 2015) implemented in RaxML, in order to correct for the use of only polymorphic sites, and found no difference between tree topologies using substitution models that did not employ this correction (data not shown). For this dataset of genomic assemblies, there appears to be no effect of using a concatenation of polymorphic sites on the resulting tree topology, although branch lengths were affected compared with an alignment including monomorphic sites.
To understand how the choice of the reference affects the analysis, NASP was also run using E. coli genome assemblies and simulated reads against the outgroup, E. fergusonii, as the reference. The results demonstrate that the same tree topology was obtained by using a different, and much more divergent, reference (compare2trees topology score =100 %). However, in both cases, fewer SNPs were identified by using a divergent reference (Table 2).
Some researchers suggest that reference-independent approaches are less biased and more reliable than reference dependent-approaches (Gardner & Hall, 2013). For the case of this E. coli dataset, the phylogeny inferred by Mugsy, a reference-independent approach, was in topological agreement with other reference-dependent approaches (Table 2). In fact, kSNPv3 was one of the only methods that returned a topology that was inconsistent with all other methods (Table 2); an inconsistent kSNP phylogeny has also been reported in the analysis of other datasets (Pettengill et al., 2014). To analyze this further, we identified SNPs (n = 826) from the NASP run using simulated paired-end reads that were uniquely shared on a branch of the phylogeny that defines a monophyletic lineage (Fig. S3). We then calculated how many of these SNPs were identified by all methods and found widely variable results (Table 2). Using kSNP with only core genome SNPs identified only five of these SNPs, which explains the differences in tree topologies.
In many cases, the same tree topology was returned even though the number of identified SNPs differed dramatically (Table 2). This result could be due to multiple factors, including if and how duplicates are filtered from the reference genome or other genome assemblies. With regards to NASP, erroneous SNPs called in genome assemblies are likely to be artifacts from the whole-genome alignments using NUCmer. The default value for aligning through poorly scoring regions before breaking an alignment in NUCmer is 200, potentially introducing spurious SNPs into the alignment, especially in misassembled regions in draft genome assemblies. By changing this value to 20, the same tree topology was obtained, although many fewer SNPs (n = approximately 100 000) were identified (Table 2). This value is easily altered in NASP and should be appropriately tuned based on the inherent expected diversity in the chosen dataset. Additional investigation is required to verify that SNPs in divergent regions are not being lost by changing this parameter. Another option is to use simulated reads from the genome assemblies in the SNP identification process.
Phylogeny differences for the same dataset
Previously, it has been demonstrated that different phylogenies can be obtained for the same dataset using either RAxML or FastTree2 (Pettengill et al., 2014). To test this result across multiple phylogenetic inference methods, the NASP E. coli read dataset was used. Phylogenies were inferred using a maximum likelihood method in RAxML, a maximum parsimony method implemented in Parsimonator, a minimum evolution method in FastTree2, and a Bayesian method implemented in Exabayes (Aberer et al., 2014). The results demonstrate variability in the placement of one genome (UMN026) depending on the method. FastTree2 and Exabayes agreed on the topology, including 100 % congruence of the replicate trees. The maximum-likelihood and maximum-parsimony phylogenies were slightly different (Fig. S3) and included low bootstrap support values at the variable node. The correct placement of UMN026 is unknown and is likely to be confounded by the extensive recombination observed in E. coli (Dykhuizen & Green, 1991).
Pipeline comparisons on a well characterized dataset
To test the functionality of different SNP calling pipelines, a set of 15 finished Y. pestis genomes were processed with NASP. This set of genomes was selected because 26 SNPs in the dataset have been verified by wet-bench methods (Table S4). Additionally, 13 known errors in the reference genome, Y. pestis CO92 (Parkhill et al., 2001), have been identified (Table S4) and should consistently be identified in SNP discovery methods. The small number of SNPs in the dataset requires accurate SNP identification to resolve the phylogenetic relationships of these genomes.
The results demonstrate differences in the total number of SNPs called between methods (Table 3). Most of the methods identified all 13 known sequencing errors in CO92, although Parsnp, REALPHY and kSNPv3 failed to do so. The number of verified SNPs identified also varied between methods, from 21 in kSNPv3 to all 26 in multiple methods (Table 3). An analysis of wet-bench-validated SNPs (n = 9) that are identified in more than one genome demonstrated that some methods failed to identify all of these SNPs, which could lead to a very different phylogeny and incorrect resolution of important phylogenetic relationships. In fact, such SNPs could represent critical markers, resulting in the inappropriate linkage or separation of strains in an outbreak event.
Table 3. SNP calling results on a set of Yersinia pestis genomes.
| Method | Data type | Parameters | Number called
SNPs |
Number CO92
errors (n=13) |
Number verified
SNPs (n=26) |
Vital SNPs
(n=9) |
|---|---|---|---|---|---|---|
| NASP | ART simulated reads | BWA, GATK, MinDepth = 3, MinAF = 0.90 | 147 | 13 | 26 | 9 |
| NASP | assemblies | default | 181 | 13 | 26 | 9 |
| ISG | ART simulated reads | minaf = 3, mindp = 0.9 | 151 | 13 | 26 | 9 |
| ISG | assemblies | default | 177 | 13 | 26 | 9 |
| Parsnp | assemblies | default | 141 | 12 | 23 | 7 |
| REALPHY | REALPHY simulated reads | default | 163 | 12 | 25 | 9 |
| SPANDx | ART simulated reads | default | 150 | 13 | 25 | 9 |
| kSNPv3 | assemblies | k=19 | 130 | 11 | 21 | 5 |
| CFSAN | ART simulated reads | default | 250 | 13 | 26 | 9 |
| lyve-SET | ART simulated reads | min_coverage 3, min_alt_frac 0.9 | 402 | 13 | 26 | 9 |
Pipeline comparisons on a simulated set of assemblies and reads
Simulated data for Y. pestis were used to compare SNP identification between pipelines. In this method, 3501 mutations (Supplemental Data File 2) were inserted into genomes based on a published phylogeny and FASTA file. Raw reads were also simulated from these artificially mutated assemblies with ART to generate paired end sequences. Reads and assemblies were run across all pipelines, where applicable.
The results demonstrate that NASP identified all of the inserted SNPs using raw reads, although 67 SNPs failed the proportion filter (0.90) and 232 SNPs fell in duplicated regions (Table 4); some of the duplicated SNPs would also fail the proportion filter. Of all other methods, only ISG identified all inserted mutations. Parsnp identified the majority of the mutations, although duplicate regions appear to have also been aligned.
Table 4. Simulated data results.
| Method | Data type | Number of called SNPs | SNPs in duplicated regions | Filtered SNPs | Total SNPs | Topological score |
|---|---|---|---|---|---|---|
| NASP | simulated reads | 3202 | 232 | 67 | 3501 | 98.50% |
| NASP | simulated assemblies | 3269 | 232 | na | 3501 | 98.50% |
| Parsnp | simulated assemblies | 3492 | unknown | na | 3492 | 95.60% |
| ISG | simulated reads | 3258 | 126 | 8 | 3392 | 92.40% |
| ISG | simulated assemblies | 3266 | 235 | na | 3501 | 95.60% |
| SPANDx | simulated reads | 3391 | unknown | 116 | 3391 | 99.20% |
| CFSAN | simulated reads | 3290 | unknown | unknown | 3290 | 95.30% |
| REALPHY | simulated assemblies | 3320 | unknown | unknown | 3320 | 91.60% |
| kSNPv3 | simulated assemblies | 3304 | unknown | na | 3304 | 91.90% |
| lyve-SET | simulated reads | 3460 | unknown | unknown | 3460 | 95.80% |
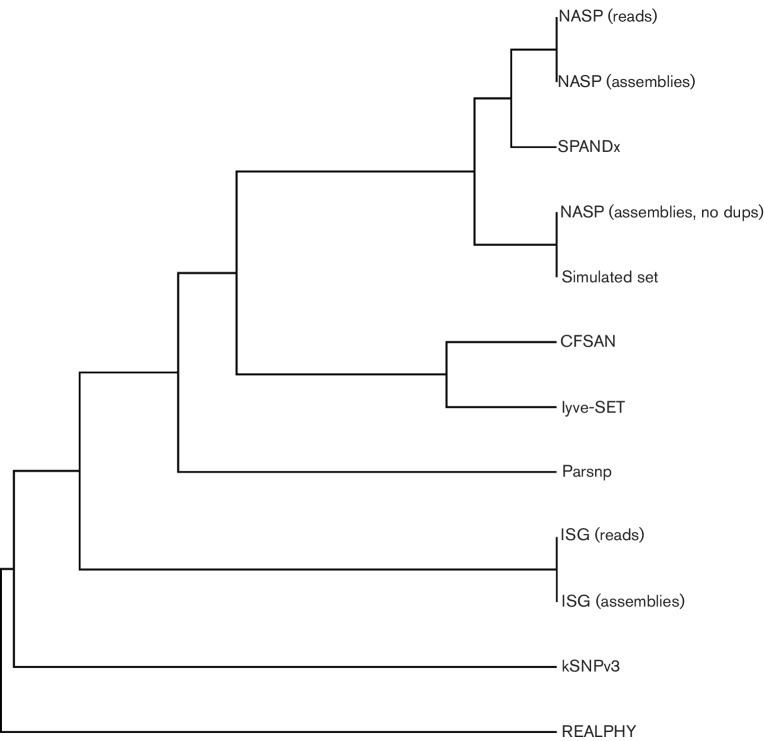
To understand how the SNPs called would affect the overall tree topology, a phylogeny was inferred for each set of SNPs with RAxML. A similarity matrix was made for each method based on the topological score compared with the ML phylogeny inferred from the known mutations. The UPGMA dendrogram demonstrates that the NASP results generally return a phylogeny that is more representative of the ‘true’ phylogeny than other methods (Fig. 3). Without removing SNPs found in duplicated regions, the NASP phylogeny was identical to the phylogeny inferred from the known SNPs.
Fig. 3.
Dendrogram of tree building methods on a simulated set of mutations in the genome of Yersinia pestis Colorado 92. The topological score was generated by compare2trees (Nye et al., 2006) compared with a maximum likelihood phylogeny inferred from a set of 3501 SNPs inserted by Tree2Reads. The dendrogram was generated with the neighbor-joining method in the Phylip software package (Felsenstein, 2005).
Comparisons between short-read aligners and SNP callers
One of the benefits of NASP is that it implements multiple SNP callers and aligners. To identify potential differences between methods, short reads simulated from the E. coli dataset were aligned against K-12 MG1655. Simulated reads were aligned against the reference genome with BWA-MEM, bowtie2 and Novoalign, and SNPs were called with the UnifiedGenotyper method in GATK. The results demonstrate that clear differences were observed between SNP sets between aligners (Fig. S4a). This demonstrates that aligners using different algorithms and alignment stringencies can differ dramatically in the set of SNPs called. To determine the effect of including a divergent outlier, the analysis was repeated excluding E. fergusonii. The results demonstrate that many more consensus SNPs were identified (79 %) between all three methods by excluding E. fergusonii compared with including the outlier (45 %) (Fig. S4a). These results indicate that some aligners are better at aligning divergent sequences using default parameters.
To test the effect of different SNP callers on the set of SNPs identified, BWA-MEM was used to align reads and SNPs were called with the UnifiedGenotyper method in GATK v2.7.2, Samtools v0.1.19 and VarScan v2.3.6. The results of this analysis demonstrated variability in the set of SNPs identified, although the variation was much less than observed between aligners (Fig. S4b). In this dataset, these small differences between SNP callers are unlikely to affect the tree topology.
To test the impact of different read aligners in a smaller dataset, reads from the Y. pestis North America dataset were aligned against CO92 and the number of called SNPs were identified. In this case, six SNPs were called that were only identified by bowtie2 and three SNPs were not identified by Novoalign. In outbreak situations where all SNPs may be needed to understand relationships, the union of SNPs called by all methods could be used. In situations where a large number of SNPs define the population structure, the intersection of all aligners and SNP callers provides high confidence, consistent calls.
Discussion
Understanding relationships between microbial isolates in a population is important for applications such as source tracking, outbreak investigations, phylogeography, population dynamics and diagnostic development. With the large number of genomes that are typically associated with these investigations, methods are required to quickly and accurately identify SNPs in a reference population. However, no studies have conducted a broad analysis to compare published methods on real and simulated datasets to identify relevant strengths and weaknesses.
Multiple publications have used a reference-dependent approach to identify SNPs to understand population dynamics. While the specific methods are often published, the pipelines to run these processes are often unpublished (den Bakker et al., 2014; Hsu et al., 2015), which complicates the ability to replicate results. NASP has already been used to identify SNPs from multiple organisms, including fungal (Engelthaler et al., 2014; Etienne et al., 2016) and bacterial (Sahl et al., 2015c; d; Bowers et al., 2015) pathogens. The version-controlled source code is available for NASP, which should ensure the replication of results across research groups.
Recently it has been suggested that the use of a single reference can bias the identification of SNPs, especially in divergent references (Bertels et al., 2014). In our E. coli test set, ~29 000 fewer SNPs were called by aligning E. coli reads against the reference genome of the outgroup, E. fergusonii, compared with the E. coli K-12 reference, although the tree topologies were identical (Table 2). In the E. coli test set phylogeny, the major clades are delineated by enough SNPs that the loss of a small percentage is insufficient to change the overall tree topology, although the branch lengths were variable. In other datasets, the choice of the reference should be made carefully to include as many SNPs as needed to define the population structure of a given dataset.
According to the authors of kSNP, a k-mer-based reference-independent approach, there are times where alignments are not appropriate in understanding bacterial population structure (Gardner & Hall, 2013). In our E. coli analysis, reference-dependent and reference-independent methods generally returned the same tree topology (Table 2), with the exception of kSNPv3 and lyve-SET, using only core genome SNPs. Using all of the SNPs identified by kSNPv3 also gave a different tree topology than the other methods (Table 2). A detailed look at branch-specific SNPs demonstrated that using kSNP with core SNPs failed to identify most of the branch-specific SNPs for one of the major defining clades (Table 2). For datasets that are only defined by a small number of SNPs, a method should be chosen that includes as many SNPs as possible in order to maximize the relevant search space. While NASP cannot truly use the pan-genome if a single reference genome is chosen, it can incorporate data from all positions in the reference genome if missing data are included in the alignment. A true pan-genome reference can be used with NASP to more comprehensively identify SNPs, but curation of the pan-genome is necessary to remove genomic elements introduced by horizontal gene transfer that could potentially confound phylogenetic inference.
Phylogenetics on an alignment of concatenated SNPs is thought to be less preferable than an alignment that also contains monomorphic positions (Bertels et al., 2014; Leache et al., 2015). However, the inclusion of monomorphic positions can drastically increase the run time needed to infer a phylogeny, especially where the population structure of a species can be determined by a small number of polymorphisms. Substitution models are available in RAxML v8 that contain acquisition bias corrections that should be considered when inferring phylogenies from concatenated SNP alignments. In our E. coli test case, using concatenated SNPs did not change the tree topology compared with a phylogeny inferred from all sites, but did affect branch lengths (Fig. S2). For downstream methods that depend on accurate branch lengths, decisions must be made on whether or not to include monomorphic positions in the alignment. NASP provides the user with the flexibility to make those decisions in a reproducible manner.
NASP represents a version-controlled, source-available, unit-tested pipeline for identifying SNPs from datasets with diverse input and output types. NASP is a high-throughput method that can take a range of input formats, can accommodate multiple job management systems, can use multiple read aligners and SNP callers, can identify both monomorphic and polymorphic sites, and can generate core genome statistics across a population.
Acknowledgements
The authors would like to acknowledge early testers of NASP for feedback and improvement suggestions.
Supplementary Data
Supplementary File 1
Abbreviations:
- SNP
Single nucleotide polymorphism
- WGS
Whole genome sequence
References
- Aberer A. J., Kobert K., Stamatakis A.(2014). ExaBayes: massively parallel Bayesian tree inference for the whole-genome era. Mol Biol Evol 312553–2556. 10.1093/molbev/msu236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiuoli S. V., Salzberg S. L.(2011). Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27334–342. 10.1093/bioinformatics/btq665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertels F., Silander O. K., Pachkov M., Rainey P. B., van Nimwegen E.(2014). Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol Biol Evol 311077–1088. 10.1093/molbev/msu088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F. R., Plunkett G., Bloch C. A., Perna N. T., Burland V., Riley M., Collado-Vides J., Rode C. K., Rode C. K., et al. (1997). The complete genome sequence of Escherichia coli K-12. Science 2771453–1462. 10.1126/science.277.5331.1453 [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B.(2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 302114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J. R., Kitchel B., Driebe E. M., MacCannell D. R., Roe C., Lemmer D., de Man T., Rasheed J. K., Engelthaler D. M., et al. (2015). Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS One 10e0133727. 10.1371/journal.pone.0133727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S. J., Lu X., Ruden D. M., Le W.(2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 680–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Yu C., Yan Y., Li D., Li Y., Jombart T., Weinert L. A., Wang Z., Guo Z., et al. (2013). Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc Natl Acad Sci U S A 110577–582. 10.1073/pnas.1205750110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher A. L., Salzberg S. L., Phillippy A. M.(2003). Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatic Chapter 10Unit 10.3. [DOI] [PubMed] [Google Scholar]
- den Bakker H. C., Allard M. W., Bopp D., Brown E. W., Fontana J., Iqbal Z., Kinney A., Limberger R., Musser K. A., et al. (2014). Rapid whole-genome sequencing for surveillance of Salmonella enterica serovar enteritidis. Emerg Infect Dis 201306–1314. 10.3201/eid2008.131399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., Hartl C., Philippakis A. A., del Angel G., Rivas M. A., et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen D. E., Green L.(1991). Recombination in Escherichia coli and the definition of biological species. J Bacteriol 1737257–7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelthaler D. M., Hicks N. D., Gillece J. D., Roe C. C., Schupp J. M., Driebe E. M., Gilgado F., Carriconde F., Trilles L., et al. (2014). Cryptococcus gattii in North American Pacific Northwest: whole-population genome analysis provides insights into species evolution and dispersal. MBio 5e01464-14–14. 10.1128/mBio.01464-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelthaler D. M., Valentine M., Bowers J., Pistole J., Driebe E. M., Terriquez J., Nienstadt L., Carroll M., Schumacher M., et al. (2016). Hypervirulent emm59 clone in invasive group A Streptococcus outbreak, southwestern United States. Emerg Infect Dis 22734–738. 10.3201/eid2204.151582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppinger M., Mammel M. K., Leclerc J. E., Ravel J., Cebula T. A.(2011). Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc Natl Acad Sci U S A 10820142–20147. 10.1073/pnas.1107176108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne K. A., Roe C. C., Smith R. M., Vallabhaneni S., Duarte C., Escadon P., Castaneda E., Gomez B. L., de Bedout C., et al. (2016). Whole-genome sequencing to determine origin of multinational outbreak of Sarocladium kiliense bloodstream infections. Emerg Infect Dis 22476–481. 10.3201/eid2203.151193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J.(2005). PHYLIP (Phylogeny Inference Package) Version 3.6, 3.6 ed University of Washington, Seattle: Department of Genome Sciences. [Google Scholar]
- Foster J. T., Beckstrom-Sternberg S. M., Pearson T., Beckstrom-Sternberg J. S., Chain P. S., Roberto F. F., Hnath J., Brettin T., Keim P.(2009). Whole-genome-based phylogeny and divergence of the genus Brucella. J Bacteriol 1912864–2870. 10.1128/JB.01581-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner S. N., Slezak T.(2010). Scalable SNP analyses of 100+ bacterial or viral genomes. J Forensic Res 01107. 10.4172/2157-7145.1000107 [DOI] [Google Scholar]
- Gardner S. N., Hall B. G.(2013). When whole-genome alignments just won't work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One 8e81760. 10.1371/journal.pone.0081760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner S. N., Slezak T., Hall B. G.(2015). kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 312877–2878. 10.1093/bioinformatics/btv271 [DOI] [PubMed] [Google Scholar]
- Hsu L. Y., Harris S. R., Chlebowicz M. A., Lindsay J. A., Koh T. H., Krishnan P., Tan T. Y., Hon P. Y., Grubb W. B., et al. (2015). Evolutionary dynamics of methicillin-resistant Staphylococcus aureus within a healthcare system. Genome Biol 1681. 10.1186/s13059-015-0643-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Li L., Myers J. R., Marth G. T.(2012). ART: a next-generation sequencing read simulator. Bioinformatics 28593–594. 10.1093/bioinformatics/btr708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L. S., Petkau A., Beaulaurier J., Tyler S., Antonova E. S., Turnsek M. A., Guo Y., Wang S., Paxinos E. E., et al. (2013). Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. MBio 4e00398-13. 10.1128/mBio.00398-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P. S., Wagner D. M.(2009). Humans and evolutionary and ecological forces shaped the phylogeography of recently emerged diseases. Nat Rev Microbiol 7813–821. 10.1038/nrmicro2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt D. C., Zhang Q., Larson D. E., Shen D., McLellan M. D., Lin L., Miller C. A., Mardis E. R., Ding L., Wilson R. K.(2012). VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22568–576. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L.(2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaché A. D., Banbury B. L., Felsenstein J., de Oca A. N., Stamatakis A.(2015). Short tree, long tree, right tree, wrong tree: new acquisition bias corrections for inferring SNP phylogenies. Syst Biol 641032–1047. 10.1093/sysbio/syv053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 252078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.(2013). Aligning sequence reads, clone sequences and assembly contigs with Bwa-Mem. arXiv.org:1303.3997 [Q-bio.Gn]. [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 201297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye T. M., Liò P., Gilks W. R.(2006). A novel algorithm and web-based tool for comparing two alternative phylogenetic trees. Bioinformatics 22117–119. 10.1093/bioinformatics/bti720 [DOI] [PubMed] [Google Scholar]
- Olson N. D., Lund S. P., Colman R. E., Foster J. T., Sahl J. W., Schupp J. M., Keim P., Morrow J. B., Salit M. L., Zook J. M.(2015). Best practices for evaluating single nucleotide variant calling methods for microbial genomics. Front Genet 6235. 10.3389/fgene.2015.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J., Wren B. W., Thomson N. R., Titball R. W., Holden M. T., Prentice M. B., Sebaihia M., James K. D., Churcher C., et al. (2001). Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413523–527. 10.1038/35097083 [DOI] [PubMed] [Google Scholar]
- Pettengill J. B., Luo Y., Davis S., Chen Y., Gonzalez-Escalona N., Ottesen A., Rand H., Allard M. W., Strain E.(2014). An evaluation of alternative methods for constructing phylogenies from whole genome sequence data: a case study with Salmonella. PeerJ 2e620. 10.7717/peerj.620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P.(2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko D. A., Webster D. R., Sahl J. W., Bashir A., Boisen N., Scheutz F., Paxinos E. E., Sebra R., Chin C. S., et al. (2011). Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med 365709–717. 10.1056/NEJMoa1106920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentería M. E., Cortes A., Medland S. E.(2013). Using PLINK for Genome-Wide Association Studies (GWAS) and data analysis. Methods Mol Biol 1019193–213. 10.1007/978-1-62703-447-0_8 [DOI] [PubMed] [Google Scholar]
- Sahl J. W., Steinsland H., Redman J. C., Angiuoli S. V., Nataro J. P., Sommerfelt H., Rasko D. A.(2011). A comparative genomic analysis of diverse clonal types of enterotoxigenic Escherichia coli reveals pathovar-specific conservation. Infect Immun 79950–960. 10.1128/IAI.00932-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl J. W., Beckstrom-Sternberg S. M., Babic-Sternberg J., Gillece J. D., Hepp C. M., Auerbach R. K., Tembe W., Wagner D. M., Keim P. S., Pearson T.(2015a). The In Silico Genotyper (ISG): an open-source pipeline to rapidly identify and annotate nucleotide variants for comparative genomics applications. bioRxiv 015578. [Google Scholar]
- Sahl J. W., Morris C. R., Emberger J., Fraser C. M., Ochieng J. B., Juma J., Fields B., Breiman R. F., Gilmour M., et al. (2015b). Defining the phylogenomics of Shigella species: a pathway to diagnostics. J Clin Microbiol 53951–960. 10.1128/JCM.03527-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl J. W., Schupp J. M., Rasko D. A., Colman R. E., Foster J. T., Keim P.(2015c). Phylogenetically typing bacterial strains from partial SNP genotypes observed from direct sequencing of clinical specimen metagenomic data. Genome Medicine 752. 10.1186/s13073-015-0176-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl J. W., Sistrunk J. R., Fraser C. M., Hine E., Baby N., Begum Y., Luo Q., Sheikh A., Qadri F., et al. (2015d). Examination of the enterotoxigenic Escherichia coli population structure during human infection. mBio 6e00501-15 10.1128/mBio.00501-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarovich D. S., Price E. P.(2014). SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets. BMC Res Notes 7618. 10.1186/1756-0500-7-618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A.(2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 301312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M., Hoede C., Tenaillon O., Barbe V., Baeriswyl S., Bidet P., Bingen E., Bonacorsi S., Bouchier C., et al. (2009). Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5e1000344. 10.1371/journal.pgen.1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen T. J., Ondov B. D., Koren S., Phillippy A. M.(2014). The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15524. 10.1186/s13059-014-0524-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharia M., Bolosky W. J., Curtis K., Fox A., Patterson D., Shenker S., Stoica I., Karp R. M., Sittler T.(2011). Faster and more accurate sequence alignment with Snap. arXiv.org: arXiv.1111.5572 [Cs.Ds] . [Google Scholar]
Data Bibliography
- Cui, Y. Sequence Read Archive. SRA010790 (2013).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1