Abstract
The DiseaseConnect (http://disease-connect.org) is a web server for analysis and visualization of a comprehensive knowledge on mechanism-based disease connectivity. The traditional disease classification system groups diseases with similar clinical symptoms and phenotypic traits. Thus, diseases with entirely different pathologies could be grouped together, leading to a similar treatment design. Such problems could be avoided if diseases were classified based on their molecular mechanisms. Connecting diseases with similar pathological mechanisms could inspire novel strategies on the effective repositioning of existing drugs and therapies. Although there have been several studies attempting to generate disease connectivity networks, they have not yet utilized the enormous and rapidly growing public repositories of disease-related omics data and literature, two primary resources capable of providing insights into disease connections at an unprecedented level of detail. Our DiseaseConnect, the first public web server, integrates comprehensive omics and literature data, including a large amount of gene expression data, Genome-Wide Association Studies catalog, and text-mined knowledge, to discover disease–disease connectivity via common molecular mechanisms. Moreover, the clinical comorbidity data and a comprehensive compilation of known drug–disease relationships are additionally utilized for advancing the understanding of the disease landscape and for facilitating the mechanism-based development of new drug treatments.
INTRODUCTION
Recent research reveals that human diseases form an interrelated landscape. Multiple diseases, even those of different organs or with distinct symptoms, can be caused by dysfunctions of the same genes, or more broadly, by dysfunction of the same pathways (1–9). For example, it is known that both asthma and type II diabetes may be linked to obesity through chronic systemic inflammation (10). In addition, many cardiovascular diseases and cancers share processes involving the endothelin axis and angiogenesis (11). However, traditional classification systems group diseases based on the similarity of their clinical presentations and phenotypic traits. Diseases with entirely different underlying pathologies could, therefore, be grouped together, leading to similar treatment designs. This problem could be avoided if diseases were classified based on their molecular mechanisms. Such a system would be a great step forward inspiring novel and effective treatment strategies of many diseases by repositioning existing drugs and therapies. To achieve this goal, a web-hosted, comprehensive database and analysis suite server on molecular-based disease mechanisms would be a very useful tool.
Recently, we and other groups have attempted to create disease connectivity networks based on clinical records (1,2), OMIM records (3,4), Genome-Wide Association Studies (GWAS) data (12), metabolic networks (5) and aggregations of these datasets (13–15). However, these studies have not yet touched two enormous and rapidly growing repositories: functional genomic data (gene expression, microRNA expression, etc.) and primary literature texts. Both resources are capable of providing insights into disease mechanisms and disease connections at an unprecedented level of detail. Currently, only a few databases touch on the concepts of disease connections (13,16). The Disease and Gene Annotation (DGA) database uses the Gene Reference Into Function (GeneRIF) from the National Center for Biotechnology Information (NCBI), Disease Ontology (DO) and molecular interaction networks to construct disease–gene, gene–gene and disease–disease relations (16). The human disease database ‘MalaCards’ archives human maladies and their annotations by integrating 44 disease- and drug-related data sources (13). As a side product, the human disease network was constructed based on shared MalaCards annotations, embodying associations based on etiology, clinical features and clinical conditions. However, none of these databases utilizes both comprehensive omics and literature data supporting shared molecular mechanisms. In addition to the disease databases, there are a number of disease–drug databases and prediction methods (17–22). For example, Sanseau et al. reported that genes with significant disease associations from GWAS studies are more likely to involve common drug targets for their respective diseases (19). Gottlieb et al. used drug–drug and disease–disease similarity to infer novel drug indications (20). The biological literature corpus has also been used to infer disease–drug relations (21,22). But these disease–drug association databases and prediction methods are independent of disease network analysis. In summary, none of this prior work focused on extensively combining and presenting a diverse range of heterogeneous resources, including genomics data, OMIM, literature, clinical data and disease–drug relations, for studying mechanism-based disease connectivity.
To fill this gap, we have recently created DiseaseConnect, a web server that focuses on the analysis of common molecular mechanisms shared by diseases by integrating comprehensive omics and literature data. Our system is equipped with efficient network analysis and visualization tools for intuitive data exploration and easy interpretation. We have already incorporated a large amount of GWAS catalog, gene expression data, microRNA expression data and text-mined knowledge to discover disease–disease connectivity based on molecular mechanisms. To advance further our understanding of the disease landscape, we have supplemented the system with clinical comorbidity data. Finally, to facilitate the development of new mechanism-based drug treatment and therapeutic strategies, we equipped the server with a comprehensive compilation of known drug–disease relationships. The DiseaseConnect web server contains 18 707 disease–disease, 660 985 disease–gene, 12 617 drug–gene and 113 498 drug–disease relations; all together, these data cover 4791 diseases, 6215 drugs and 15 182 genes. This is, therefore the most comprehensive resource documenting the shared molecular bases of diseases currently in existence.
The web server uses the advanced web interactive visualization technology ‘Cytoscape Web’ (23), which provides an interactive and user-friendly interface to visualize networks and results in many different ways. Nodes, edges and labels of different types of networks are rendered with distinct and easy-to-recognize colors and sizes. The interface also supports various network layouts, zooming and draggable functions to facilitate network exploration and comprehension of the analysis and results.
To validate the server's usefulness to biologists, we show that diseases with shared molecular mechanisms are likely (i) to be linked with clinical comorbidity and (ii) to have the common drug treatments. Through several examples, we also demonstrate how DiseaseConnect can be used to reveal the molecular mechanisms shared between diseases and suggest potential drug treatments. In summary, our DiseaseConnect web server characterizes common pathobiological mechanisms across diseases in different organ systems. This new and rapidly expanding tool has the great potential to redefine disorders based on their underlying molecular and cellular pathobiology. It is also useful for the rational, mechanism-based development of new diagnostic, prognostic and therapeutic strategies.
MATERIALS AND METHODS
Our web server compiles a set of comprehensive data resources that can be categorized into eight types: disease annotations, disease-related gene and microRNA expression changes, disease-related single-nucleotide polymorphisms (SNPs), disease–drug relationships, disease comorbidity relationships, drug–gene relations, disease–gene relationships from literature mining and disease–gene relationships from OMIM. The processed data are organized by the system schema shown in Figure 1. This section details the processing procedure for each data type.
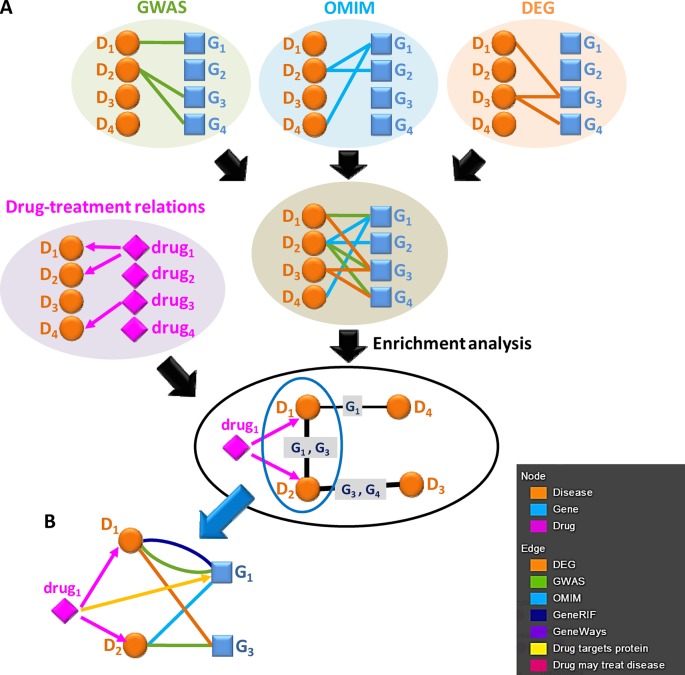
Figure 1.
Constructing the mechanism-based disease–disease network based on the GWAS/OMIM/DEG records. (A) We combine the disease–gene connections derived from GWAS, OMIM and DEG records to build a comprehensive disease–gene network (D1, D2, D3 and D4 indicate diseases and G1, G2, G3 and G4 indicate genes). For each disease pair, we calculate the hypergeometric P-value to assess the significance of the number of genes involved in both diseases. We also add drug–treatment relations to complement further the database. (B) In the disease–disease network, when users click the edge between disease D1 and D2, the web server generates the detailed network of disease D1 and D2, including DEG/GWAS/OMIM/GeneRIF/GeneWays disease–gene relations and drug treatment/target relations.
Disease annotations
To construct systematically disease–gene and disease–drug relations using multiple databases, we used the unified medical language system (UMLS) that provides a comprehensive set of medical concepts and DO terms and includes a metathesaurus (24). We selected two vocabularies: Medical Subject Headings (MeSH) and the UMLS Metathesaurus. To describe disease–gene relations, DiseaseConnect uses only UMLS concepts with one of the following disease-related semantic types: ‘Pathologic Function’, ‘Injury or Poisoning’ and ‘Anatomical Abnormality’. To support a wide range of diseases, we combined all of the disease concepts from gene expression data, OMIM and GWAS databases. To avoid too general concepts, we only used those UMLS concepts that are associated with < 50 descendant concepts. These criteria result in a universe of 4791 disease concepts.
Disease-related gene expression
We collected a total of 1366 human gene expression datasets on 27 February 2014 from the Gene Expression Omnibus (GEO) (25), which consist of all 1355 human GEO datasets (GDS) that are pre-processed by GEO, and additionally 11 GEO series (GSE) processed by us as part of an NIH-funded project studying heart, lung, blood and sleep disorders. If one gene has several probes, we used the mean expression value for the gene. Within each sample, the expression values of all genes were log-transformed, median-centered and normalized to have unit standard deviation. The 1366 datasets contain in total 6024 subsets and 30 300 samples. Each subset is a group of samples with specific phenotypic traits or treatments. These subsets can be classified into different types, e.g. disease state, agent, cell line, cell type, tissue, protocol and infection. To concentrate on the disease studies, we selected 1178 subsets with the ‘disease state’ type. We manually assigned UMLS concepts to subsets, and then retrieved their parental UMLS concepts to complete the UMLS annotation for all subsets. To identify differentially expressed genes (DEGs) in each dataset, we selected all disease subsets with at least three samples and manually identified the normal subset in the dataset. We then performed a t-test between two subsets without overlapping samples. DEGs are defined as the top 100 genes with t-test P-values < 1e−6 for each subset pair. This analysis identified 928 subset pairs with DEGs. Overall, the 1366 gene expression datasets contributed 173 992 disease–gene relations, including 505 diseases and 11 812 genes.
Disease-related microRNA expression
We collected 233 human microRNA expression datasets from the GEO, which consist of 5194 samples. Each dataset has multiple phenotype-specific subsets. For studying disease-related microRNA data, we selected 84 datasets with disease and normal subsets. To systematically process phenotype information for microRNA expression data, we transferred the text description of microRNA datasets to UMLS concepts using the MetaMap program (26). To identify differentially expressed microRNAs in each dataset, we selected the disease subsets with at least three samples as well as the normal subset. We then performed a t-test between the normal subset and each disease subset without overlapping samples. We obtained the differentially expressed microRNAs with t-test P-values < 0.001 for each subset pair. This analysis resulted in 956 differentially expressed microRNAs identified in 73 subset pairs. The microRNA expression data contributed 17 088 disease–microRNA relations, including 20 diseases and 745 microRNAs.
Disease-related SNP
We downloaded the catalog of published GWAS from the National Human Genome Research Institute (27). This data is a manual curation of published GWAS hits, which may contain causative SNPs (27). The GWAS catalog contains descriptions of diseases/traits, associated SNPs, and reported genes. We converted these disease annotations to UMLS concepts using the MetaMap program (26) and selected GWAS genes with P-values < 1e−6. The GWAS data contributed 21 865 disease–gene relations, including 719 diseases and 3397 genes.
Disease–drug relationships
We extracted the disease–drug annotations and relationships from the UMLS National Drug File Reference Terminology (NDFRT). This database contributed 113 498 drug–disease relations, including 8995 drugs and 1525 diseases.
Disease comorbidity relationships
We downloaded disease comorbidity relations from the Phenotypic Disease Network (PDN) (1), which uses ICD9 disease codes. We selected disease–disease connections with t-value > 1.96 (P < 0.05), and then translated the ICD9 disease codes into UMLS concepts using the UMLS Metathesaurus. The PDN data contributed 132 786 disease–disease connections, including 2991 diseases.
Drug–gene relations
We downloaded 12 617 human drug–gene relations from the DrugBank database (28). These data included 3417 drugs and 1481 genes.
Disease–gene relationships from GeneRIF
GeneRIF is a database archiving short statements about the functions and disease relevance of genes. Each GeneRIF statement has a pointer to the PubMed ID of a scientific publication that provides evidence for the statement. We downloaded the GeneRIF database from the NCBI Gene database, and then for each gene we extracted UMLS disease concepts from its GeneRIF statements using the UMLS natural language processing tool, MetaMap program (26). This source provided 276 089 disease–gene relations, including 3113 diseases and 11 190 genes.
Disease–gene relationships from literature corpus mining (GeneWays)
We extracted all human disease–gene relationships from the GeneWays system (version 8.0) that we developed previously (29). This system has automatically extracted, analyzed, visualized and integrated molecular pathway data in 483 107 full-text articles and 11 826 299 abstracts downloaded from PubMed. We obtained 170 329 disease–gene relations, including 2046 diseases and 7239 genes. This set of disease–gene relationships has less than 20% overlap (33 137 shared) with those derived from GeneRIF. This difference in the two derived knowledge sources can be attributed to the different text-mining approaches: GeneRIF was curated by a mixed effort of manual annotation and various user-designed text-mining approaches, while GeneWays used natural language processing tools. The two sets of disease–gene relationships are complementary to a great extent; we, therefore, retained both resources for our web server.
Disease–gene relationships from OMIM
We downloaded the OMIM database (30), then converted annotations of the OMIM Gene Map to UMLS concepts using the UMLS natural language processing tool (26). This analysis resulted in 18 710 disease–gene relations, including 4457 diseases and 3749 genes.
After collecting the above data, we constructed a disease–disease connectivity network summarizing the molecules shared between diseases. We evaluated the connection strength between two diseases based on the enrichment of genes that are relevant to both diseases. The statistical significance of the connection between two diseases is assessed by a hypergeometric test on shared genes derived from the GWAS, OMIM, DEG, GeneRIF and GeneWays sources. We did not include disease–disease connections derived from GeneRIF and GeneWays in this server, because the disease–gene relationships extracted from the curated literature have low performance in terms of the relatedness to the comorbidity and drug treatment data (see Figures 3 and 4). In addition, those literature-based disease–gene relationships were included in the server as the complementary data.
Figure 3.

Diseases pairs sharing more involved genes are more likely to have disease comorbidity. The statistical significance (P-value) of the connection between two diseases is assessed by a hypergeometric test on shared genes derived from each individual data source of GWAS, OMIM, DEG, GeneRIF and GeneWays. We used various P-value thresholds (x axis) to select significant disease–disease connections for each data source, and then calculated the fraction (y axis) of those disease–disease connections that overlap with the disease comorbidity connections.
Figure 4.

Disease pairs sharing more genes are more likely to have the same drug treatment. The statistical significance (P-value) of the connection between two diseases is assessed by a hypergeometric test on shared genes derived from each individual data source of GWAS, OMIM, DEG, GeneRIF and GeneWays. We used various P-value thresholds (x axis) to select significant disease–disease connections for each data source, and then calculated the fraction (y axis) of the disease–disease connections in which both diseases can be treated by the same drug(s).
SERVER IMPLEMENTATION AND USAGE
The DiseaseConnect web server was implemented using JSP, MySQL, JavaScript and an advanced web interactive visualization technology, Cytoscape Web (23). The server permits rapid, dynamic user interaction and network visualization. The nodes, edges and labels of different types of networks are rendered with distinct and easy-to-recognize colors and sizes. Users can choose between various network layouts, zoom in and out and use draggable functions to facilitate better understanding of the analysis and results.
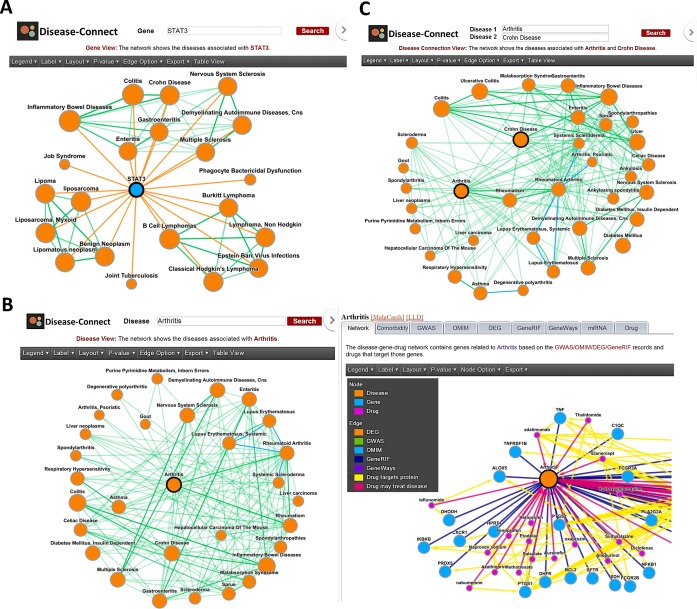
As illustrated in Figure 2, the server provides three views: a gene view, a disease view and a disease connection view. In the gene view (Figure 2A), users enter a gene of interest, and the web server returns a set of diseases that have similar molecular mechanisms as well as association with the queried gene. The diseases are, therefore, represented as networks, where edges indicate diseases with a shared molecular basis. The strength of the connection is quantified as the P-value of a hypergeometric enrichment test in the number of shared genes; the P-value threshold is set by the server to limit the size of the network to 150 nodes. Users can also enter a disease (in the disease view, Figure 2B) or a disease pair (in the disease connection view, Figure 2C). In both cases, the web server returns all diseases connected to the queried disease(s) in a network representation. Again, the edges in these networks indicate a strong connection in terms of shared genes, as explained for the gene view.
Figure 2.
Illustrations of the gene, disease and disease connection views in the DiseaseConnect web server. (A) Gene view: user inputs STAT3 to search for all diseases related to this gene based on the GWAS/OMIM/DEG records. The web server generates a disease–disease network on the diseases associated with STAT3. In this network, the disease–disease connection indicates that two diseases involve a significant number of common genes based on the GWAS/OMIM/DEG records. (B) Disease view: user inputs arthritis, and then the web server displays a disease–disease network (using the diseases connecting arthritis) in the left panel and the disease–gene–drug network (using the arthritis-related drugs and genes) in the right panel. The disease–gene–drug network of arthritis includes the following molecules: (i) genes associated with arthritis, (ii) the drugs targeting the disease-related genes and (iii) the drugs treating arthritis. (C) Disease connection view: user inputs arthritis and Crohn disease, and then the web server displays a network connecting the two diseases. Users can enable the drug option to show the drug–disease treatment relations. In each view, the web server automatically adjusts the P-value threshold to maintain the size of the network to less than 100 nodes.
The interface includes numerous features to facilitate exploration for the user. If the user enters only the starting symbol of a gene or disease, the auto-complete field automatically provides a list of partially matched terms. To retrieve all genes and drugs associated with a disease or connection of interest, the users can click a node or edge of the disease network. The web server then retrieves and displays the gene–drug network associated with the disease or disease connection of interest. This network includes seven types of associations: DEG, GWAS, OMIM, GeneRIF, GeneWays, Drug target and Drug treatment. For detailed query examples, refer to the section ‘Example Applications’.
In summary, the main features of the DiseaseConnect web server are as follows:
Construct comprehensive networks describing disease–disease connectivity, disease–gene associations, drug–gene targeting and drug–disease treatments.
Integrate detailed lists and representations of disease–gene relations derived from various sources (GWAS, OMIM, DEG, GeneRIF and GeneWays).
Provide flexible network visualization tools with different choices of network layout, as well as customizable node and edge types; allow the user to zoom and drag the network diagram.
Use edge opacity to indicate the strength of the disease–disease connections in the network.
Automatically adjust the hypergeometric P-value threshold to present a subnetwork of a reasonable size.
Auto-complete the search field with full names of diseases, UMLS IDs and gene symbols.
Export the network as standard image file formats, such as PNG, SVG and XGMML.
VALIDATION
In this section, we theoretically validate the utility of our server in helping biologists study mechanism-based disease connectivity. The following analyses show that diseases with shared molecular mechanisms are likely (i) to be linked with clinical comorbidity data and (ii) to have the same drug treatment.
Diseases with shared molecular mechanisms are more likely to have clinical comorbidity
We assessed the strength of disease connections based on the significance of the number of genes involved in both disorders, which were derived from genomic data and the literature. We are interested in the extent to which these mechanism-based disease connections are related to disease comorbidity relationships of the same disease pairs, based on patient clinical records. The mechanism-based and comorbidity-based disease networks can be compared in terms of the fraction of shared edges. To construct the comorbidity network, we used only high-quality comorbidity relationships, with P-values <0.01, retrieved from the PDN (1). So that the comparison is unbiased, we excluded disease pairs with ancestor–descendant relationships in the disease semantic hierarchy (i.e. ‘is-a’ relations of UMLS disease concepts). The results of this comparison are shown in Figure 3. The overall trend is that two diseases with stronger mechanism-based connections are more likely to have a significant clinical comorbidity relation. When we randomize the mechanism-based disease network by degree-preserving edge rewiring (31) , this trend disappears and the random network shares few edges with the disease comorbidity network.
Among all data sources, the disease network derived from GWAS records showed the highest level of consistency with the comorbidity network. Interestingly, the OMIM-derived network not only is less consistent but has the ‘seesaw’ performance curve. The DEG- and GWAS-based networks both have stable performance. A possible explanation is that although OMIM has high-quality data and has expanded to polygenic complex maladies in recent years (4,32), many (i.e. 35.5%) of its records are monogenic disorders for which our assessment of disease connection strength may not be precise. Finally, it is not surprising that the networks based on GeneRIF and GeneWays data from the literature have the worst performance because they include too much noisy data incurred by the use of automatic human language processing tools. However, both these networks still perform better than chance. Overall, these results validate our claim that diseases with shared molecular mechanisms are more likely to have comorbidity.
Diseases with shared molecular mechanisms are more likely to have the same drug treatment
To validate the intuitive hypothesis that diseases with shared molecular mechanisms are more likely to be treated by the same drug(s), we performed a network comparison similar to that described above. In this case, we calculated the fraction of mechanism-based disease connections for which both diseases are treated by the same drug or drugs. The drug–disease treatment resources were collected from NDFRT (see the Material and Methods section). We individually evaluated the disease networks derived from different sources (GWAS/OMIM/DEG/GeneWays/GeneRIF). For an unbiased comparison, we used only those diseases with drug–treatment relations and excluded disease pairs with ancestor–descendant relationships in the disease semantic hierarchy (i.e. ‘is-a’ relations of UMLS disease concepts).
Figure 4 displays the results of this comparison: (i) the more significant the mechanism-based connection between two diseases (derived from any data sources), the higher the likelihood they can be treated by the same drugs. For example, for the data source OMIM, we identified 1243 disease–disease connections (covering 424 diseases and 599 drugs) in each of which both diseases share significant (P-value <0.01) common molecular mechanisms and both diseases have known drug–treatment data. Among those, the fraction in which both diseases are treated by the same drug(s) for OMIM is 33%. Figure 4 shows that this fraction increases with the strength (i.e. P-value) of mechanism-based disease connections. (ii) Connections derived from the GWAS and OMIM data sources are the strongest predictors of common drug treatments. It is also interesting to note that while connections derived from text mining (GeneRIF/GeneWays) are not good predictors, they still perform better than random networks. The GWAS data source consistently outperforms OMIM by a small amount. The chance for two diseases to be treated by the same drug(s) increases with the significance of their GWAS- or OMIM-based connection; however, DEG-based connections show no such trend. This implies that the molecular mechanisms indicated by GWAS and OMIM records are more closely relevant to drug treatment than those present in the DEG records. A possible reason is that DEG records, derived from gene expression data, may more likely reflect downstream effect rather than causes of diseases.
Diseases modules in the connectivity network are more likely to have the same drug treatment
Having confirmed that disease pairs with significant mechanism-based connections are likely to be treated by the same drug(s), we further hypothesize that diseases comprising a dense subgraph in the disease connectivity network, termed a disease module (33), are likely to benefit from the same drug treatment. To validate this hypothesis, we constructed a large, high-quality disease connectivity network, comprising 5189 connections with hypergeometric P-values < 1e-6 for disease module discovery. We applied MODES, a network clustering method to discover overlapping dense clusters (34) in this network and identified 141 distinct disease modules (details see Supplementary Table S1). We ran MODES using the following parameters: minimum module size 3, maximum module size 30 and density cutoff 0.7. In 38 (27%) of the discovered modules, more than half of the member diseases can be treated by the same drug. In contrast, our randomization test (we generated 141 similar-sized modules by randomly permuting drug–treatment relations) has only 3 (2.1%) such modules. This result indicates that highly connected diseases are more likely to have the same drug treatment. Figure 5 illustrates Module-109 as an example. This densely connected disease subnetwork consists of nine diseases, of which the drug ‘fludarabine’ can treat five. Fludarabine is effective in the treatment of chronic lymphocytic leukemia (35). Although multiple myeloma (MM) and acute lymphocytic leukemia (ALL) are not currently treated with fludarabine, we found strong evidence for a potential effect on MM and ALL in the literature: (i) Caballero-Velázquez et al. reported that a combined treatment consisting of bortezomib, fludarabine, and melphalan showed advantage in high-risk MM patients (36). (ii) Daly et al. reported that fludarabine and busulfan achieved excellent outcomes with older ALL patients (37). (iii) There are several ongoing clinical trials using fludarabine to treat MM (e.g. ClinicalTrials.gov ID: NCT01131169 and NCT01453101) and ALL (e.g. ClinicalTrials.gov ID: NCT01435447 and NCT01572662). Therefore, identifying disease modules such as this one in the disease connectivity network can point the way to effective drug repositioning strategies.
Figure 5.

Disease module 109 is enriched of the diseases that can be treated by fludarabine. There are nine total diseases, and fludarabine can treat five of those diseases. Although multiple myeloma and acute lymphocytic leukemia are currently not treated with fludarabine, we found strong literature evidence that supports this potential treatment.
EXAMPLE APPLICATIONS
DiseaseConnect offers a variety of functions and analyses. While a mechanism-based disease network has many potential applications, this section provides examples for two basic and yet informative functions of the server, e.g. how to query a disease for obtaining its related diseases, genes and drugs, and how best to query a disease–disease connection for exploring possible drug repositioning.
Example of a disease query (arthritis)
When researching a disease, biologists usually consider what other diseases are relevant to it. In the disease view, users can enter a full or partial disease name in the input field. The server automatically matches and completes a partial name with a list of full disease names. For example, when entering ‘arthritis’, the web server yields the disease–disease network shown in the left panel of Figure 2B. In this network, a disease–disease connection indicates that two diseases involve a significant number of shared genes based on the GWAS/OMIM/DEG records. If the user clicks a disease (node) in the network, the web server shows the network of all molecules associated with this disease. For example, the right panel of Figure 2B shows the disease–gene–drug network associated with arthritis, which includes the following molecules: (i) genes associated with arthritis based on the GWAS/OMIM/DEG/GeneRIF/GeneWays records, (ii) drugs that target these genes and (iii) drugs that treat arthritis.
Given the heterogeneous diseases connected to arthritis in Figure 2B, it is worth noting that arthritis is already known to relate to large group diseases, subsuming its Diagnosis Related Group (DRG). DRG is a system to classify hospital cases into one of originally 467 groups (38) and is currently the main system documenting disease groups. DRG however has major shortcomings being highly focused and directly linked to specific molecular mechanisms in some cases (such as mitochondriopathies or muscular dystrophies) and highly nonspecific with a clinical syndrome emphasis in others (such as heart failure). Our system, however, provides a comprehensive and unbiased way to identify diseases sharing molecular similarities, given a large amount of collected genomic and literature data.
Examples of disease pair queries and their applications to revealing potential drug treatments
After viewing all diseases and molecules related to a query disease, users can further explore the shared molecular mechanism(s) between two specific diseases by clicking an edge in the disease network. For example, when a user queries MM, the web server generates a disease–disease network. The user can then click the connection between MM and hemorrhagic disorders (HD) to view a network of shared genes and drugs between HD and MM. Figure 6A shows a small part of this network with the shared gene PSMB5 and the drug bortezomib. PSMB5 is a DEG for both HD and MM, and also has a GeneRIF association with MM. Bortezomib is a therapeutic proteasome inhibitor for treating MM, and targets PSMB5 (39). Interestingly, bortezomib can also treat HD (40), suggesting that shared disease genes can serve as good targets for drug repositioning strategies.
Figure 6.

Disease connections with drug treatment implications. (A) PSMB5 is a DEG for both hemorrhagic disorders and multiple myeloma, and PSMB5 also has GeneRIF association with multiple myeloma. (B) Arthritis and Crohn disease are associated with TNF based on the GWAS and GeneRIF records. Thalidomide is an immunomodulatory drug that targets TNF and can treat both arthritis and Crohn diseases. Adalimumab also targets TNF and can treat arthritis, suggesting its potential treatment of Crohn disease.
Figure 6B shows another example network for the queried disease pair ‘arthritis’ and ‘Crohn disease’, both involve the tumor necrosis factor (TNF), based on the data sources GWAS and GeneRIF. Thalidomide is an immunomodulatory drug that targets TNF. Interestingly, thalidomide can treat both arthritis and Crohn diseases (41), supporting our hypothesis that diseases with a shared molecular mechanism are likely to be treated by the same drug. Another drug shown in Figure 6B, ‘adalimumab’, is known to target TNF and can treat arthritis, suggesting that it may also be effective for Crohn disease. In fact, this inference is confirmed independently by a recent report of Peters et al. (42). Also, several clinical trials about Adalimumab and Crohn disease (e.g. ClinicalTrials.gov ID: NCT01556672 and NCT01958827) are currently ongoing.
CONCLUSIONS
We developed the DiseaseConnect web server for the analysis and visualization of a comprehensive knowledge based on shared molecular mechanisms between diseases, including gene expression data, GWAS hits, OMIM records, text mining of the literature, clinical comorbidity data and a comprehensive compilation of known drug–disease relationships. Our analyses have shown that disease connections based on a shared molecular mechanism are closely tied to disease comorbidity and common drug treatments. The web server will not only facilitate studies of disease mechanisms but also has practical usages for the mechanism-based development of new drug treatments and therapeutic strategies. The latter were demonstrated by two online analysis examples. DiseaseConnect possesses an interactive and user-friendly visualization interface powered by advanced web interactive visualization technology. Therefore, it has a quick response time and an intuitive schema so that both novice and experienced users can readily perform a variety of network analyses. With the rapid accumulation of disease-related omics data and text literature that are publicly available, our server will be continuously updated to serve the research and clinical communities effectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be considered as Joint First Authors.
FUNDING
The National Institutes of Health (NIH) [NHLBI MAPGEN U01HL108634 and NIGMS R01GM105431] and the National Science Foundation [0747475 to X.J.Z.]; the National Science Council grant [NSC101–2627-B-005–002] and the Ministry of Education, Taiwan, R.O.C. under the ATU plan (to C.C.L.); the National Institutes of Health (NIH) [NHLBI MAPGEN U01HL108630 to J.L.]. Funding for open access charge: NHLBI MAPGEN U01HL108634.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hidalgo C.A., Blumm N., Barabási A.-L., Christakis N.A. A dynamic network approach for the study of human phenotypes. PLoS Comput. Biol. 2009;5:e1000353. doi: 10.1371/journal.pcbi.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rzhetsky A., Wajngurt D., Park N., Zheng T. Probing genetic overlap among complex human phenotypes. Proc. Natl Acad. Sci. U.S.A. 2007;104:11694–11699. doi: 10.1073/pnas.0704820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Driel M.A., Bruggeman J., Vriend G., Brunner H.G., Leunissen J.A.M. A text-mining analysis of the human phenome. Eur. J. Hum. Genet. 2006;14:535–542. doi: 10.1038/sj.ejhg.5201585. [DOI] [PubMed] [Google Scholar]
- 4.Goh K.-I., Cusick M.E., Valle D., Childs B., Vidal M., Barabási A.-L. The human disease network. Proc. Natl Acad. Sci. U.S.A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee D.-S., Park J., Kay K.A., Christakis N.A., Oltvai Z.N., Barabási A.-L. The implications of human metabolic network topology for disease comorbidity. Proc. Natl Acad. Sci. U.S.A. 2008;105:9880–9885. doi: 10.1073/pnas.0802208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linghu B., Snitkin E.S., Hu Z., Xia Y., Delisi C. Genome-wide prioritization of disease genes and identification of disease-disease associations from an integrated human functional linkage network. Genome Biol. 2009;10:R91. doi: 10.1186/gb-2009-10-9-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oti M., Brunner H.G. The modular nature of genetic diseases. Clin. Genet. 2007;71:1–11. doi: 10.1111/j.1399-0004.2006.00708.x. [DOI] [PubMed] [Google Scholar]
- 8.Oti M., Huynen M.A., Brunner H.G. Phenome connections. Trends Genet. 2008;24:103–106. doi: 10.1016/j.tig.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Loscalzo J., Kohane I., Barabasi A.-L. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol. Syst. Biol. 2007;3:124. doi: 10.1038/msb4100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sin D.D., Sutherland E.R. Obesity and the lung: 4. Obesity and asthma. Thorax. 2008;63:1018–1023. doi: 10.1136/thx.2007.086819. [DOI] [PubMed] [Google Scholar]
- 11.Minotti G., Salvatorelli E., Menna P. Pharmacological foundations of cardio-oncology. J. Pharmacol. Exp. Ther. 2010;334:2–8. doi: 10.1124/jpet.110.165860. [DOI] [PubMed] [Google Scholar]
- 12.Barrenas F., Chavali S., Holme P., Mobini R., Benson M. Network properties of complex human disease genes identified through genome-wide association studies. PLoS One. 2009;4:e8090. doi: 10.1371/journal.pone.0008090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rappaport N., Nativ N., Stelzer G., Twik M., Guan-Golan Y., Stein T.I., Bahir I., Belinky F., Morrey C.P., Safran M., et al. MalaCards: an integrated compendium for diseases and their annotation. Database (Oxford). 2013;2013:bat018. doi: 10.1093/database/bat018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zitnik M., Janjić V., Larminie C., Zupan B., Pržulj N. Discovering disease-disease associations by fusing systems-level molecular data. Sci. Rep. 2013;3:3202. doi: 10.1038/srep03202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis D.A., Chawla N. V. Exploring and exploiting disease interactions from multi-relational gene and phenotype networks. PLoS One. 2011;6:e22670. doi: 10.1371/journal.pone.0022670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng K., Xu W., Zheng J., Huang K., Wang H., Tong J., Lin Z., Liu J., Cheng W., Fu D., et al. The Disease and Gene Annotations (DGA): an annotation resource for human disease. Nucleic Acids Res. 2013;41:D553–560. doi: 10.1093/nar/gks1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., Lerner J., Brunet J.-P., Subramanian A., Ross K.N., et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 18.Chiang A.P., Butte A.J. Systematic evaluation of drug-disease relationships to identify leads for novel drug uses. Clin. Pharmacol. Ther. 2009;86:507–510. doi: 10.1038/clpt.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanseau P., Agarwal P., Barnes M.R., Pastinen T., Richards J.B., Cardon L.R., Mooser V. Use of genome-wide association studies for drug repositioning. Nat. Biotechnol. 2012;30:317–320. doi: 10.1038/nbt.2151. [DOI] [PubMed] [Google Scholar]
- 20.Gottlieb A., Stein G.Y., Ruppin E., Sharan R. PREDICT: a method for inferring novel drug indications with application to personalized medicine. Mol. Syst. Biol. 2011;7:496. doi: 10.1038/msb.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frijters R., van Vugt M., Smeets R., van Schaik R., de Vlieg J., Alkema W. Literature mining for the discovery of hidden connections between drugs, genes and diseases. PLoS Comput. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheung W.A., Ouellette B.F.F., Wasserman W.W. Compensating for literature annotation bias when predicting novel drug-disease relationships through Medical Subject Heading Over-representation Profile (MeSHOP) similarity. BMC Med. Genomics. 2013;6 Suppl 2:S3. doi: 10.1186/1755-8794-6-S2-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopes C.T., Franz M., Kazi F., Donaldson S.L., Morris Q., Bader G.D. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aronson A.R., Lang F.-M. An overview of MetaMap: historical perspective and recent advances. J. Am. Med. Inform. Assoc. 2010;17:229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knox C., Law V., Jewison T., Liu P., Ly S., Frolkis A., Pon A., Banco K., Mak C., Neveu V., et al. DrugBank 3.0: a comprehensive resource for “omics” research on drugs. Nucleic Acids Res. 2011;39:D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rzhetsky A., Iossifov I., Koike T., Krauthammer M., Kra P., Morris M., Yu H., Duboué P.A., Weng W., Wilbur W.J., et al. GeneWays: a system for extracting, analyzing, visualizing, and integrating molecular pathway data. J. Biomed. Inform. 2004;37:43–53. doi: 10.1016/j.jbi.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 30. OMIM - Online Mendelian Inheritance in Man.
- 31.Maslov S., Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- 32.Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barabási A.-L., Gulbahce N., Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu H., Yan X., Huang Y., Han J., Zhou X.J. Mining coherent dense subgraphs across massive biological networks for functional discovery. Bioinformatics. 2005;21(Suppl. 1):i213–221. doi: 10.1093/bioinformatics/bti1049. [DOI] [PubMed] [Google Scholar]
- 35.Rai K.R., Peterson B.L., Appelbaum F.R., Kolitz J., Elias L., Shepherd L., Hines J., Threatte G.A., Larson R.A., Cheson B.D., et al. Fludarabine compared with chlorambucil as primary therapy for chronic lymphocytic leukemia. N. Engl. J. Med. 2000;343:1750–1757. doi: 10.1056/NEJM200012143432402. [DOI] [PubMed] [Google Scholar]
- 36.Caballero-Velázquez T., López-Corral L., Encinas C., Castilla-Llorente C., Martino R., Rosiñol L., Sampol A., Caballero D., Serrano D., Heras I., et al. Phase II clinical trial for the evaluation of bortezomib within the reduced intensity conditioning regimen (RIC) and post-allogeneic transplantation for high-risk myeloma patients. Br. J. Haematol. 2013;162:474–482. doi: 10.1111/bjh.12410. [DOI] [PubMed] [Google Scholar]
- 37.Daly A., Savoie M.L., Geddes M., Chaudhry A., Stewart D., Duggan P., Bahlis N., Storek J., Brown C., Shafey M., et al. Fludarabine, busulfan, antithymocyte globulin, and total body irradiation for pretransplantation conditioning in acute lymphoblastic leukemia: excellent outcomes in all but older patients with comorbidities. Biol. Blood Marrow Transplant. 2012;18:1921–1926. doi: 10.1016/j.bbmt.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 38.Fetter R.B., Shin Y., Freeman J.L., Averill R.F., Thompson J.D. Case mix definition by diagnosis-related groups. Med. Care. 1980;18:1–53. iii. [PubMed] [Google Scholar]
- 39.Ri M., Iida S., Nakashima T., Miyazaki H., Mori F., Ito A., Inagaki A., Kusumoto S., Ishida T., Komatsu H., et al. Bortezomib-resistant myeloma cell lines: a role for mutated PSMB5 in preventing the accumulation of unfolded proteins and fatal ER stress. Leukemia. 2010;24:1506–1512. doi: 10.1038/leu.2010.137. [DOI] [PubMed] [Google Scholar]
- 40.Sinn D.-I., Lee S.-T., Chu K., Jung K.-H., Kim E.-H., Kim J.-M., Park D.-K., Song E.-C., Kim B.-S., Yoon S.-S., et al. Proteasomal inhibition in intracerebral hemorrhage: neuroprotective and anti-inflammatory effects of bortezomib. Neurosci. Res. 2007;58:12–18. doi: 10.1016/j.neures.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 41.Franks M.E., Macpherson G.R., Figg W.D. Thalidomide. Lancet. 2004;363:1802–1811. doi: 10.1016/S0140-6736(04)16308-3. [DOI] [PubMed] [Google Scholar]
- 42.Peters C.P., Eshuis E.J., Toxopeüs F.M., Hellemons M.E., Jansen J.M., D’Haens G.R.A.M., Fockens P., Stokkers P.C.F., Tuynman H.A.R.E., van Bodegraven A.A. Adalimumab for Crohn's disease: Long-term sustained benefit in a population-based cohort of 438 patients. J. Crohns. Colitis. 2014 doi: 10.1016/j.crohns.2014.01.012. doi:10.1016/j.crohns.2014.01.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.