Abstract
Parkinson's disease (PD) is a disorder of movement, cognition, and emotion, and it is characterized pathologically by neuronal degeneration with Lewy bodies, which are cytoplasmic inclusion bodies containing deposits of aggregated proteins. Most PD cases appear to be sporadic, but genetic forms of the disease, caused by mutations in α-synuclein, parkin, and other genes, have helped elucidate pathogenesis. Mutations in leucine-rich repeat kinase 2 (LRRK2) cause autosomal-dominant Parkinsonism with clinical features of PD and with pleomorphic pathology including deposits of aggregated protein. To study expression and interactions of LRRK2, we synthesized cDNAs and generated expression constructs coding for human WT and mutant LRRK2 proteins. Expression of full-length LRRK2 in cells in culture suggests that the protein is predominately cytoplasmic, as is endogenous protein by subcellular fractionation. Using coimmunoprecipitation, we find that LRRK2, expressed in cells in culture, interacts with parkin but not with α-synuclein, DJ-1, or tau. A small proportion of the cells overexpressing LRRK2 contain protein aggregates, and this proportion is greatly increased by coexpression of parkin. In addition, parkin increases ubiquitination of aggregated protein. Also, mutant LRRK2 causes neuronal degeneration in both SH-SY5Y cells and primary neurons. This cell model may be useful for studies of PD cellular pathogenesis and therapeutics. These findings suggest a gain-of-function mechanism in the pathogenesis of LRRK2-linked PD and suggest that LRRK2 may be involved in a pathogenic pathway with other PD-related proteins such as parkin, which may help illuminate both familial and sporadic PD.
Keywords: cell death, Parkinson's disease, protein aggregation, protein interaction, ubiquitin
Parkinson's disease (PD) is a progressive neurodegenerative disorder characterized clinically by tremor, rigidity, and bradykinesia and pathologically by loss of dopamine neurons in substantia nigra and other brain regions (1-4). There are characteristic ubiquitinated inclusions (proteinaceous aggregates), termed Lewy bodies. In PD and related disorders, aggregates are located in the substantia nigra and other brain regions (1, 4, 5). The pathogenesis of PD remains incompletely understood, but it appears to involve both genetic susceptibility and environmental factors. Genes whose mutations have been shown to cause Parkinsonism with features comparable with PD include α-synuclein, parkin, DJ-1, and PINK1 (6-13).
Mutations in the leucine-rich repeat kinase 2 (LRRK2) gene can cause autosomal-dominant PD (14-16). The gene product has also been termed “Dardarin.” Several different point mutations segregate with the PD phenotype in families, including the original PARK8 family (17-22), and they are found also in some sporadic PD cases (23). Patients with LRRK2 mutations have typical relatively late onset Parkinsonism with features comparable with idiopathic PD, with asymmetric rest tremor, bradykinesia, rigidity, and a good response to 3,4-dihyroxy-l-phenylalanine (l-DOPA) (15, 24). The pathology of cases with LRRK2 mutations is pleomorphic (15, 17). There are typical features of neuronal loss and gliosis in the substantia nigra. In some cases, there are Lewy bodies and Lewy neurites, sometimes located diffusely in the brain. In other cases, there are protein deposits, mostly in the cytoplasm, with some heterogeneity both within and between families.
LRRK2 is a protein with 2,527 aa, including the following several predicted functional domains: Roc (Ras in complex proteins), COR domain (C-terminal of Roc), a leucine-rich repeat consisting of 12 repetitions of a 22- to 28-aa motif, a protein kinase catalytic domain [mitogen-activated protein kinase kinase kinase (MAPKKK), described as a tyrosine kinase], a WD40 domain, and an ankryin domain (14, 15, 25). Point mutations have been found in almost all of the identified domains. The distribution of mutations in several different domains, as well as the lack of deletions or truncations, along with dominant inheritance, are consistent with a gain-of-function mechanism.
In this study, we investigated the cellular localization of LRRK2 and its possible interactions with other PD-related gene products, and we sought to determine whether LRRK2 may be involved in cell toxicity. We found that LRRK2 had a predominantly cytoplasmic localization. LRRK2 interacted with parkin but not α-synuclein, DJ-1, or tau. Mutant LRRK2 dramatically decreased cell viability in culture with either SH-SY5Y cells or primary neurons. These findings contribute to the understanding of LRRK2-linked PD pathogenesis.
Materials and Methods
Materials. Media, N2 and N27 supplements for cell culture, Lipofectamine Plus reagent, and anti-hemagglutinin (HA) polyclonal Ab were obtained from Invitrogen. Hoechst 33342 was obtained from Molecular Probes. Anti-ubiquitin polyclonal Ab was obtained from DAKO. Anti-myc and anti-HA mAbs were obtained from Santa Cruz Biotechnology and Roche Molecular Biochemicals. CyTM3-conjugated goat anti-mouse IgG and FITC-conjugated goat anti-rabbit IgG were obtained from Jackson ImmunoResearch. Anti-FLAG Ab was obtained from Sigma. Anti-histone 2B and anti-tubulin Abs were obtained from Upstate Biotechnology (Lake Placid, NY).
LRRK2 cDNA Synthesis, Plasmids, and Transfection. WT and mutant LRRK2 cDNAs were synthesized by DNA 2.0, Inc. (Menlo Park, CA) according to the published sequence (GenBank accession no. AY792511; ref. 15). We added three FLAG tags to the N terminus of LRRK2 cDNA and inserted the construct into the pcDNA3.1 vector. We also generated the version of C-terminal myc-tagged WT LRRK2 construct for coimmunoprecipitation experiments. We constructed a series of FLAG-tagged truncated LRRK2 plasmids using WT LRRK2 as a template by PCR. All resulting constructs were confirmed by sequencing. Plasmids expressing HA-α-synuclein, HA-parkin, myc-parkin, FLAG-parkin, and full-length huntingtin (htt) with 23 polyglutamine (Q) have been described (26-28). Transient transfections were performed with Lipofectamine Plus according to the manufacturer's protocol.
Cell Cultures, Immunoprecipitation (IP), and Western Blot Analysis. Human HEK 293T and SH-SY5Y cells were grown in the media as described (29, 30). We synthesized two peptides to generate Abs against two distinct regions of human LRRK2 (amino acids 1245-1259 and 2396-2408). Cells were harvested in the lysis buffer as described in ref. 31. The resulting lysates were subjected to Bradford protein assay to ensure equal protein loading. IP experiments from transfected cell lysates were performed with anti-FLAG, or anti-HA Ab and protein G Plus/protein A-agarose (Amersham Pharmacia Biotech). The resulting immunoprecipitates and cell lysates were resolved on 4-12% NuPAGE Bis-Tris gels and transferred onto polyvinylidene difluoride membranes (Invitrogen). The membranes were blocked in TBST (10 mM Tris·HCl, pH 7.4/150 mM NaCl/0.1% Tween 20) containing 5% nonfat milk and then probed with different Abs. Proteins were detected by using enhanced chemiluminescence reagents (NEN).
Subcellular Fractionation of Endogenous LRRK2 in SH-SY5Y Cells. SH-SY5Y cells were harvested in lysis buffer containing 0.25 M sucrose, 25 mM KCl, 5 mM MgCl2, 50 mM triethanolamine, 1 mM PMSF, 1 mM DTT, and complete protease inhibitors. Cells were incubated on ice for 30 min, and complete lysis of the plasma membrane was visualized by using 2× trypan blue solution containing Hoechst 33342. Lysates were centrifuged at 800 × g for 15 min to generate supernatant (cytosol) and crude pellet (containing nuclear) fractions. The pellet was purified further by the sucrose cushion buffer (lysis buffer with 1.8 M sucrose) and centrifugation at 30,000 × g for 1 h. Protein content was determined by using the BCA (bicinchoninic acid) protein assay kit (Pierce).
Mouse Primary Cortical Neuronal Cultures and Electroporation Transfection. Mouse primary cortical neuronal cultures were derived from CD-1 outbred mice (The Jackson Laboratory) at embryonic day 15 or 16. Cortices were dissociated as described in ref. 29. After dissociation, neurons were plated on laminin- and poly-d-lysine-coated plates (BD Biosource, San Diego) and cultured in neurobasal medium with the addition of Glutamax, B-27 supplement, and penicillin/streptomycin. Under these culture conditions, 95% of cells were neurons. Transfection of LRRK2 constructs into mouse primary cortical neurons was carried out by using Nucleofector (Amaxa Biosystem, Cologne, Germany), which is an electroporation-based method, as described in ref. 30.
Measurement of Cell Viability and Cell Death. SH-SY5Y cell viability assays were conducted as described in ref. 29. Cells were cotransfected with pcDNA3.1-GFP, along with vector pcDNA3.1, full-length htt with 23Q, and pcDNA3.1-FLAG-LRRK2 constructs (at 1:15) for 24 h in 10% FBS OPTI-I media and then changed to DMEM with N2 supplement for 24 h. Primary neurons were cotransfected with LRRK2 constructs or full-length htt with 23Q, and a plasmid encoding GFP (at 15:1) for 48 h. GFP-positive viable cells (neurons) were counted from 40 randomly selected fields by an investigator, who was kept unaware of the experimental condition, using fluorescence microscopy. Viable cells (neurons) were defined as having at least one smooth extension (neurite) with twice the length of the cell body. The percentage of GFP-positive viable cells (neurons) in each experimental group relative to those of cells (neurons) transfected with vector and GFP was calculated.
Hoechst 33342/propidium iodide labeling of cells to detect apoptotic and necrotic cell death was performed as described in ref. 29. TUNEL staining was performed as described (30) by using the Texas red in situ cell-death-detection kit (Roche Molecular Biochemicals). SH-SY5Y cells were cotransfected with various constructs for 24 h in 10% FBS OPTI-I media, changed to DMEM with N2 supplement for 24 h, and then subjected anti-FLAG immunochemical staining. TUNEL staining was then performed according to the manufacturer's instructions. For counting, images were taken from 20 randomly selected fields from each experimental group by using conventional fluorescence microscopy by an investigator who was kept unaware of the experimental condition.
Immunocytochemistry. Cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and processed as described (31). Cell preparations were incubated with primary Abs: anti-FLAG, anti-myc, or anti-ubiquitin, then with secondary Abs: Cy3-conjugated anti-mouse or FITC-conjugated anti-rabbit; nuclei were stained with Hoechst 33342, and all signals were analyzed by fluorescence and confocal microscopy (LSM 510 and Axiovert 100, Zeiss). The number of cells with cytoplasmic aggregates were counted in 20 randomly selected fields with ≈1,000 cells in each experimental condition. Counts were done by an investigator who was kept unaware of the experimental condition.
Data Analysis. Quantitative data are expressed as arithmetic means ± SE based on at least three separate experiments performed in duplicate. The difference between two groups was statistically analyzed by Student's t test or one-way ANOVA. Significance was defined at P < 0.05.
Results
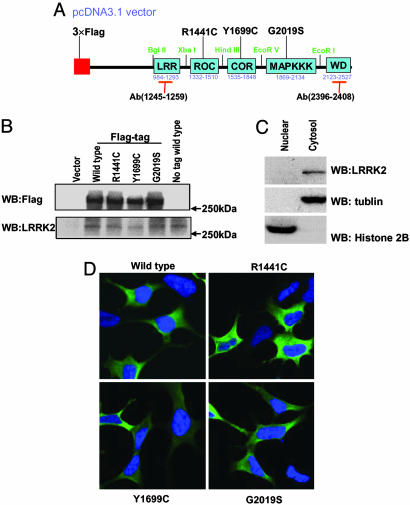
Cytoplasmic Expression of LRRK2. To study the expression of LRRK2, we generated a cDNA expression vector containing the full-length LRRK2 ORF with codons optimized for mammalian and bacterial expression (Fig. 1A). Several versions of the construct were generated, including full-length WT LRRK2 with no tag and constructs with N-terminal FLAG tag or C-terminal myc tag. Also, the following three of the reported mutants were generated: R1441C in the ROC domain, Y1699C in the COR domain, and G2019S (the most common one) in the mitogen-activated protein kinase kinase kinase (MAPKKK) domain, with FLAG or myc tags. We synthesized two peptides to generate Abs against two distinct regions of human LRRK2 (amino acids 1245-1259 and 2396-2408).
Fig. 1.
Cytoplasmic expression of LRRK2. (A) Schematic representation of LRRK2 expression construct in pcDNA3.1 vector. The WT construct was designed with or without a FLAG tag. Unique restriction sites were engineered between domains to facilitate cloning. (B) Lysates derived from HEK 293T cells transiently expressing various LRRK2 constructs or empty vector were analyzed by Western blotting using anti-FLAG and anti-LRRK2-(1245-1259) Abs. LRRK2 proteins migrated at ≈280 kDa. Similar results were obtained by using anti-LRRK2-(2396-2408) Ab (data not shown). (C) Subcellular fractionations derived from SH-SY5Y cells were analyzed by Western blot analysis using anti-LRRK2-(1245-1259) Abs to detect the endogenous LRRK2. (D) Representative merged confocal images of transiently transfected HEK 293T cells. Nuclear Hoechst 33342 staining is indicated blue, and green indicates FLAG-FITC Ab staining for LRRK2.
Expression of LRRK2 in HEK 293T cells yielded a protein of ≈280 kDa detected with either Abs to the FLAG tag or to LRRK2 (Fig. 1B). The WT LRRK2 protein without FLAG-tag ran at a similar position (Fig. 1B). Cellular expression of WT or mutant LRRK2 appeared predominately diffuse cytoplasmic (Fig. 1D), although a minority of cells had a punctate label suggestive of aggregates (see below). There was no apparent change in the cytoplasmic distribution for any of the mutants in HEK 293T cells. Subcellular fractionations derived from SH-SY5Y cells confirmed that LRRK2 was excluded from the nucleus and present in cytosol (Fig. 1C).
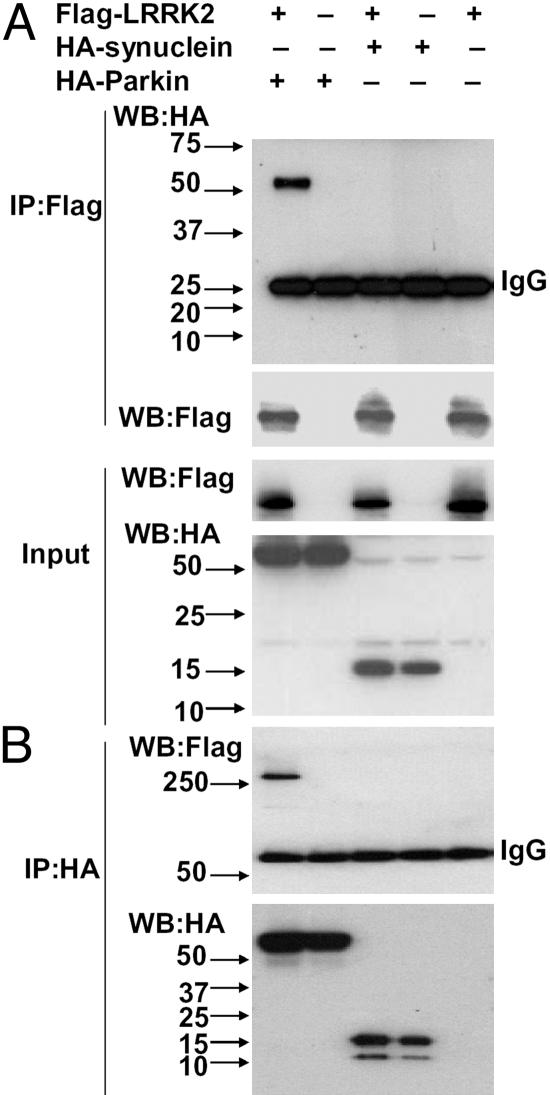
Interaction of LRRK2 with Parkin. Because LRRK2 showed predominately cytoplasmic expression, we investigated whether it interacted with other cytoplasmic PD-related gene products. By coimmunoprecipitation assays, LRRK2 showed a specific interaction with parkin but not with α-synuclein (Fig. 2), DJ-1, or tau (data not shown). Fig. 2A shows IP with FLAG-LRRK2 and detection of either HA-α-synuclein or HA-parkin. Parkin coimmunoprecipitated with LRRK2, but α-synuclein did not. Conversely, either α-synuclein or parkin was immunoprecipitated by using HA, followed by anti-FLAG LRRK2 immunoblotting (Fig. 2B). LRRK2 was detected only with IP of HA-parkin. Even when the blot was considerably overexposed, there was no detection of LRRK2 using IP of HA-α-synuclein (data not shown). The LRRK2 mutations did not alter the interaction between LRRK2 and parkin (data not shown).
Fig. 2.
LRRK2 interaction with parkin in HEK 293T cells. (A and B) Lysates prepared from cells transfected with various constructs as indicated were subjected to IP with anti-FLAG (A) or anti-HA (B), followed by anti-HA and anti-FLAG immunoblotting. The experiment was repeated three times with similar results.
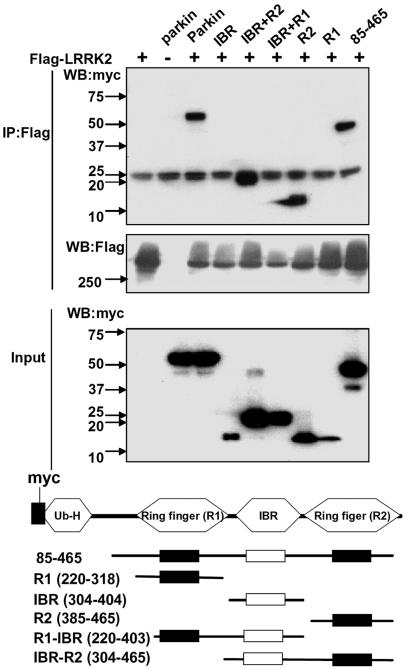
To determine which regions of parkin and LRRK2 were responsible for the interaction, a series of constructs containing different domains of parkin tagged with myc were cotransfected with full-length LRRK2. As shown in Fig. 3, only parkin constructs containing the RING2 domain showed an interaction with LRRK2. Conversely, when different domains of FLAG-LRRK2 were cotransfected with full-length HA-parkin, there was a strong interaction between the COR domain of LRRK2 and parkin (data not shown).
Fig. 3.
LRRK2 interacts preferentially with the C-terminal R2 ring-finger domain of parkin. Lysates prepared from HEK 293T cells cotransfected with FLAG-LRRK2 and various myc-tagged parkin domain constructs were subjected to IP with anti-FLAG, followed by anti-myc immunoblotting. Putative functional domains of parkin used in the mapping experiments are shown. Ub-H, Ubiquitin homology domain. Cell lysates were subject to Bradford protein assay to ensure equal protein loading. Parkin fragments had variable levels, even when we loaded the same amount of total protein, presumably because these nonphysiological forms are not stable. On a qualitative basis, the RING2 domain of parkin interacts most strongly with LRRK2
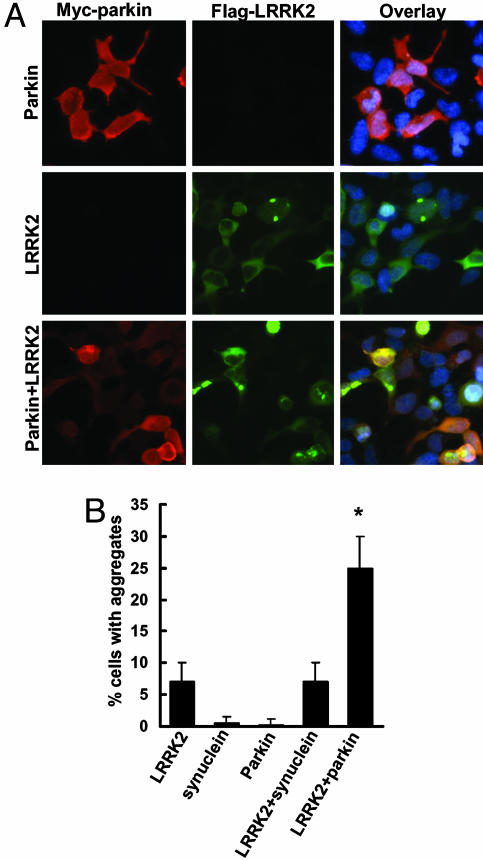
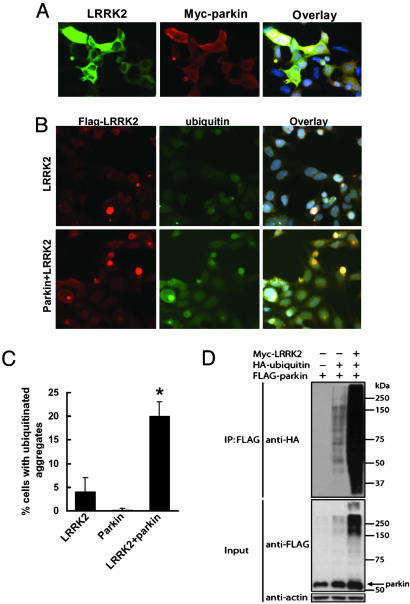
Increase of Cytoplasmic Aggregates Containing LRRK2 with Coexpression of Parkin. Although the predominant localization of LRRK2 in transfected cells is diffusely cytoplasmic, a minority of cells show apparent aggregates (Fig. 4A). There was no consistent change in the percentage of cells with aggregates using the mutants (data not shown). However, when LRRK2 was cotransfected with parkin, there was a substantial increase in the percentage of cells with aggregates (Fig. 4). By contrast, there was no change detected when LRRK2 was transfected with α-synuclein or full-length htt with 23Q.
Fig. 4.
Expression of parkin increases cytoplasmic aggregates containing LRRK2. (A and B) Cells were transfected with WT FLAG-LRRK2 with or without myc-parkin for 72 h and subjected to immunocytochemical assay with anti-myc and anti-FLAG Abs. (A) Representative images of each experimental group. (B) Cells with cytoplasmic aggregates containing FLAG-LRRK2 labeling were counted. Data are shown as means ± SE for three separate experiments performed in duplicate. *, P < 0.05 vs. cells transfected with FLAG-LRRK2 alone.
Because parkin is an E3 ubiquitin ligase (27, 32-34), we sought to determine whether the aggregates in these cells contain ubiquitinated proteins. As shown in Fig. 5 B and C, there was a considerable increase in the percentage of cells containing ubiquitinated aggregates when LRRK2 was cotransfected with parkin. We also found that ≈5% of cells with aggregates contained both parkin and LRRK2 labeling (Fig. 5A). We did not find evidence that LRRK2 might be directly ubiquitinated by parkin (data not shown). In cotransfection experiments, LRRK2 caused a 25-fold increase in the autoubiquitination activity of parkin in the context of a 3-fold increase in parkin protein level (Fig. 5D).
Fig. 5.
Expression of parkin increases ubiquitinated cytoplasmic aggregates containing LRRK2. (A and B) Cells were transfected with WT FLAG-LRRK2 with or without myc-parkin for 72 h and subjected to immunocytochemical assay with anti-FLAG and anti-myc (A) or anti-FLAG and anti-ubiquitin (B) Abs. (C) Cells with cytoplasmic aggregates containing both ubiquitin and FLAG labeling were counted. Data are shown as means ± SE for three separate experiments performed in duplicate. *, P < 0.05 vs. cells transfected with FLAG-LRRK2 alone. (D) LRRK2 enhances parkin autoubiquitination. Cells cotransfected with FLAG-parkin, HA-ubiquitin, myc-LRRK2, or EGFP plasmid as a control for 48 h and then were subjected to IP with anti-FLAG Ab. Immunoprecipitates were analyzed by Western blotting with anti-HA Ab, and inputs were probed with anti-FLAG or anti-actin Abs. Note that LRRK2 enhances the formation of high-molecular-weight parkin-ubiquitin protein conjugates corresponding to polyubiquitinated forms of parkin. These experiments were replicated three times with similar results.
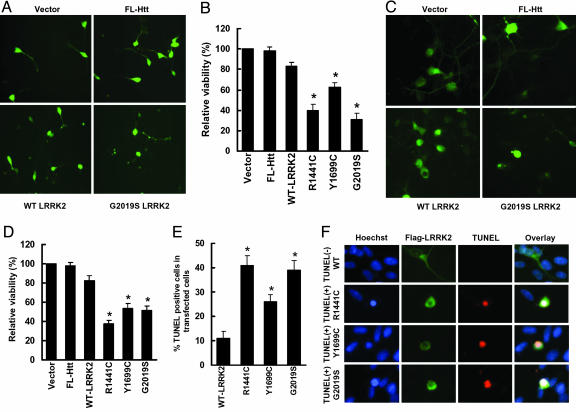
Neuronal Degeneration Caused by Expression of Mutant LRRK2. To determine whether mutant LRRK2 could alter cell viability, SH-SY5Y cells were cotransfected with GFP and various LRRK2 constructs by Lipofectamine. Viable cells were defined as having at least one smooth extension with twice the length of the cell body, and they were counted by an investigator who was kept unaware of the experimental condition. Transfection efficiencies were similar in all groups. There was mild, nonsignificant, decreased viability with WT LRRK2. All three mutations of LRRK2 caused significant cell toxicity compared with WT LRRK2, control protein (full-length htt with 23Q), and vector (Fig. 6 A and B). We confirmed these results using mouse primary cortical neurons and electroporation. Mutant LRRK2 caused significant neuronal degeneration in primary cultures compared with WT LRRK2, control protein (full-length htt with 23Q), and vector (Fig. 6 C and D). Expression of mutant LRRK2 in SH-SY5Y cells caused highly condensed and fragmented nuclei as measured by Hoechst 33342/propidium iodide staining and significantly increased the TUNEL-positive cells (Fig. 6 E and F), indicating that mutant LRRK2 induced primarily apoptotic cell death. Coexpression of parkin did not significantly protect against mutant LRRK2-induced neuronal degeneration (data not shown).
Fig. 6.
Mutant LRRK2 causes neuronal degeneration. (A) SH-SY5Y cells were cotransfected with pcDNA3.1-GFP along with either vector pcDNA3.1, full-length htt with 23 Q (FL-Htt), WT, or mutant pcDNA3.1-FLAG-LRRK2 at 1:15 ratios for 24 h in 10% FBS OPTI-MEM I media, then incubated with DMEM containing N2 supplement for 24 h. GFP-positive cells with 2-fold continuous extensions were counted by using fluorescence microscopy. Shown are representative photomicrographs for each experimental group. Transfection efficiency of GFP was ≈10%. (B) Quantitation of data in A, representing the cell viability of each experimental group normalized to that of cells cotransfected with empty vector and GFP. The percentage of GFP-positive viable cells in each experimental group relative to those of cells cotransfected with vector and GFP was calculated. *, P < 0.05 vs. cells cotransfected with WT LRRK2 and GFP. (C) Mouse primary cortical neurons were cotransfected with pcDNA3.1-GFP along with various constructs as in A at 1:15 for 48 h by electroporation. GFP-positive viable neurons with 2-fold continuous neurites were counted by using fluorescence microscopy. Shown are representative photomicrographs for each experimental group. (D) Quantitation of data in C, representing the cell viability of each experimental group normalized to that of cells cotransfected with empty vector and GFP as in B. *, P < 0.05 vs. cells cotransfected with WT LRRK2 and GFP. (E and F) SH-SY5Y cells were cotransfected with various constructs for 24 h in 10% FBS OPTI-MEM I media and then incubated with DMEM containing N2 supplement for 24 h, followed by anti-FLAG immunostaining and TUNEL assays. (F) Representative confocal images of transiently transfected SH-SY5Y cells. Blue indicates nuclear Hoechst 33342 staining, green indicates FLAG-FITC Ab staining for LRRK2, and red indicates TUNEL staining. (E) Quantitation of data in F, representing the percentage of TUNEL-positive cells in total LRRK2 transfected cells. *, P < 0.05 vs. cells transfected with WT LRRK2.
Discussion
In this study, we found that LRRK2 protein was predominately cytoplasmic. Using coimmunoprecipitation, we found that LRRK2 interacted with parkin, but not with α-synuclein, DJ-1, or tau. LRRK2 interacted preferentially with the C-terminal R2 RING-finger domain of parkin, and parkin interacted with the COR domain of LRRK2. Coexpression of LRRK2 and parkin increased cytoplasmic protein aggregates that contain LRRK2 and enhanced the ubiquitination of these aggregates. Last, expression of mutant LRRK2 caused neuronal degeneration in both SH-SY5Y cells and mouse primary neurons. These findings suggest a gain-of-function mechanism in the pathogenesis of LRRK2-linked disease.
The cytoplasmic localization of LRRK2 protein is similar to that of several other PD-related gene products, including α-synuclein and parkin (35, 36). LRRK2 appears to be expressed in most tissues although at low levels (14). The cytoplasmic localization is consistent with the LRRK2 amino acid sequence, which does not contain any predicted hydrophobic membrane spanning domains or targeting sequences for other cellular organelles (15). However, we cannot exclude the possibility that some fraction of LRRK2 protein might be associated with cytoskeleton or with the cytoplasmic faces of mitochondrial, endoplasmic reticulum, or other membranes.
We found that LRRK2 specifically associated with parkin but not α-synuclein, DJ-1, or tau protein. Parkin is an E3 ubiquitin ligase, and several of its substrates and interaction partners are relevant for PD pathogenesis (11, 27, 37-39). Parkin associates with and ubiquitinates synphilin-1 (27), a protein that interacts with α-synuclein (28) and is highly enriched in Lewy bodies (40). Also, parkin can interact with other proteins, such as p38/JTv-1 (38).
The site of interaction between LRRK2 and parkin appears to be in the RING2 domain of parkin. This site is also the site of parkin interaction with synphilin-1 (27). When expressed in cells, LRRK2 had a tendency to aggregate, and aggregates were strikingly increased when LRRK2 was coexpressed with parkin but not other proteins. We also found that cytoplasmic aggregates were ubiquitinated in a parkin-dependent fashion. Previously, we found that parkin promotes the formation of ubiquitinated aggregates with cotransfected α-synuclein and synphilin-1 (31). Other studies also found that parkin can accumulate in aggresomes under conditions of proteasome impairment (41, 42). These observations are consistent with the idea of a role for the ubiquitin proteasome pathway in PD (43, 44). In this study, the effect of parkin on the ubiquitination of aggregates may involve, in part, stimulation of the ubiquitin ligase activity of parkin by LRRK2. Preliminary data (data not shown) suggest LRRK2 can be ubiquitinated but not by parkin. Thus, parkin may ubiquitinate other proteins within the aggregates. Patients with LRRK2 mutations have pleomorphic brain pathology, including several different kinds of protein aggregates, suggesting that our findings in cell culture may be relevant to LRRK2-linked pathogenesis. It is not yet known whether LRRK2 protein is contained in these aggregates in PD patients. A caveat to this study is that most of the data were obtained by using overexpression of exogenous proteins and would need to be extended to endogenous proteins when reagents are available. LRRK2 appears to be expressed at relatively low levels in brain, and these studies would require development of Abs that are sufficiently sensitive and specific to detect such low levels of protein.
We found that mutant LRRK2 strikingly decreased neuronal cell viability in both SH-SY5Y cells and primary neurons and caused apoptotic cell death in SH-SY5Y cells. In previous experiments using comparable systems, we have shown highly specific cell toxicity caused by mutant htt, but not normal htt, indicating that this kind of transient transfection cell model can give specific disease-related results (26, 30). WT LRRK2 also caused some mild decreased cell viability. Endogenous WT LRRK2 could be involved in toxicity under some circumstances, particularly when combined with exogenous cellular stressors, raising the possibility that these effects could be relevant for sporadic PD as well. We did not find any strong evidence for parkin to protect against LRRK2 toxicity, consistent with recent studies suggesting that the role of parkin in the formation of Lewy bodies and cellular protection may be independent (36, 45, 46).
PD is similar to other neurodegenerative diseases with both protein aggregation and neuronal cell death, although the relation between aggregation and cell death may be complex (5, 47, 48). The study of α-synuclein has provided insight into both genetic and sporadic PD (49). Similarly, the study of LRRK2 cell biology has the potential to identify the contribution of LRRK2 to neuronal cell death and aggregate formation, and to the molecular pathogenesis of both familial and sporadic PD, which may suggest approaches to therapeutic development. For PD, like Alzheimer's disease, the identification of rare genetic forms has led to the identification of critical gene products that are likely to be involved in the pathogenesis of sporadic disease. Protein interactions are believed to be central to Alzheimer's disease pathogenesis. For example, the presenilin gene product has a key role in the proteolysis of amyloid precursor protein (APP), yielding the toxic amyloid-β peptide (50-52).
Our results indicate that LRRK2 is a cytoplasmic protein and that mutant LRRK2 can be directly toxic to cells, although these observations would need to be extended to endogenous protein in vivo and human tissue studies. The cell model reported here may be useful for studies of PD pathogenesis and, possibly, therapeutics. LRRK2 interacted with parkin, and parkin can potentiate the formation of aggregates containing LRRK2 and ubiquitin. These findings may be relevant to the formation of Lewy bodies or other protein aggregates in PD and related disorders. Identification of LRRK2 interaction partners and toxicity is likely to provide insights into the molecular pathway of PD pathogenesis and the targets for therapeutic intervention.
Acknowledgments
We thank Dr. Akira Sawa for helpful discussions and Brad Harris for technical help. This work was supported by National Institute of Neurological Disorders and Stroke Grants NS38377, NS054817, and 16375; the Udall PD Research Center; the National Institutes of Health; the National Parkinson's Foundation; and the American Parkinson's Disease Association. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases.
Author contributions: W.W.S., Z.P., V.L.D., T.M.D., and C.A.R. designed research; W.W.S., Z.P., H.J., D.J.M., Y.L., and A.B.W. performed research; W.W.S. analyzed data; and W.W.S. and C.A.R. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PD, Parkinson's disease; LRRK2, leucine-rich repeat kinase 2; IP, immunoprecipitation; Htt, huntingtin; HA, hemagglutinin; Q, polyglutamine.
References
- 1.Dauer, W. & Przedborski, S. (2003) Neuron 39, 889-909. [DOI] [PubMed] [Google Scholar]
- 2.Forno, L. S. (1996) J. Neuropathol. Exp. Neurol. 55, 259-272. [DOI] [PubMed] [Google Scholar]
- 3.Mouradian, M. M. (2002) Neurology 58, 179-185. [DOI] [PubMed] [Google Scholar]
- 4.Dawson, T. M. & Dawson, V. L. (2003) Science 302, 819-822. [DOI] [PubMed] [Google Scholar]
- 5.Taylor, J. P., Hardy, J. & Fischbeck, K. H. (2002) Science 296, 1991-1995. [DOI] [PubMed] [Google Scholar]
- 6.Kruger, R., Kuhn, W., Muller, T., Woitalla, D., Graeber, M., Kosel, S., Przuntek, H., Epplen, J. T., Schols, L. & Riess, O. (1998) Nat. Genet. 18, 106-108. [DOI] [PubMed] [Google Scholar]
- 7.Polymeropoulos, M. H., Lavedan, C., Leroy, E., Ide, S. E., Dehejia, A., Dutra, A., Pike, B., Root, H., Rubenstein, J., Boyer, R., et al. (1997) Science 276, 2045-2047. [DOI] [PubMed] [Google Scholar]
- 8.Zarranz, J. J., Alegre, J., Gomez-Esteban, J. C., Lezcano, E., Ros, R., Ampuero, I., Vidal, L., Hoenicka, J., Rodriguez, O., Atares, B., et al. (2004) Ann. Neurol. 55, 164-173. [DOI] [PubMed] [Google Scholar]
- 9.Hattori, N., Kitada, T., Matsumine, H., Asakawa, S., Yamamura, Y., Yoshino, H., Kobayashi, T., Yokochi, M., Wang, M., Yoritaka, A., et al. (1998) Ann. Neurol. 44, 935-941. [DOI] [PubMed] [Google Scholar]
- 10.Kitada, T., Asakawa, S., Hattori, N., Matsumine, H., Yamamura, Y., Minoshima, S., Yokochi, M., Mizuno, Y. & Shimizu, N. (1998) Nature 392, 605-608. [DOI] [PubMed] [Google Scholar]
- 11.Moore, D. J., Zhang, L., Troncoso, J., Lee, M. K., Hattori, N., Mizuno, Y., Dawson, T. M. & Dawson, V. L. (2005) Hum. Mol. Genet. 14, 71-84. [DOI] [PubMed] [Google Scholar]
- 12.Bonifati, V., Dekker, M. C., Vanacore, N., Fabbrini, G., Squitieri, F., Marconi, R., Antonini, A., Brustenghi, P., Dalla, L. A., De Mari, M. et al. (2002) Neurol. Sci. 23, Suppl. 2, S59-S60. [DOI] [PubMed] [Google Scholar]
- 13.Moore, D. J., West, A. B., Dawson, V. L. & Dawson, T. M. (2005) Annu. Rev. Neurosci. 28, 57-87. [DOI] [PubMed] [Google Scholar]
- 14.Paisan-Ruiz, C., Jain, S., Evans, E. W., Gilks, W. P., Simon, J., van der, B. M., de Munain, A. L., Aparicio, S., Gil, A. M., Khan, N., et al. (2004) Neuron 44, 595-600. [DOI] [PubMed] [Google Scholar]
- 15.Zimprich, A., Biskup, S., Leitner, P., Lichtner, P., Farrer, M., Lincoln, S., Kachergus, J., Hulihan, M., Uitti, R. J., Calne, D. B., et al. (2004) Neuron 44, 601-607. [DOI] [PubMed] [Google Scholar]
- 16.Shen, J. (2004) Neuron 44, 575-577. [DOI] [PubMed] [Google Scholar]
- 17.Funayama, M., Hasegawa, K., Ohta, E., Kawashima, N., Komiyama, M., Kowa, H., Tsuji, S. & Obata, F. (2005) Ann. Neurol. 57, 918-921. [DOI] [PubMed] [Google Scholar]
- 18.Lewthwaite, A. J. & Nicholl, D. J. (2005) Curr. Neurol. Neurosci. Rep. 5, 397-404. [DOI] [PubMed] [Google Scholar]
- 19.Morris, H. R. (2005) Ann. Med. 37, 86-96. [DOI] [PubMed] [Google Scholar]
- 20.Gasser, T. (2005) Curr. Opin. Neurol. 18, 363-369. [DOI] [PubMed] [Google Scholar]
- 21.Farrer, M., Stone, J., Mata, I. F., Lincoln, S., Kachergus, J., Hulihan, M., Strain, K. J. & Maraganore, D. M. (2005) Neurology 65, 738-740. [DOI] [PubMed] [Google Scholar]
- 22.Tan, E. K., Shen, H., Tan, L. C., Farrer, M., Yew, K., Chua, E., Jamora, R. D., Puvan, K., Puong, K. Y., Zhao, Y., et al. (2005) Neurosci. Lett. 384, 327-329. [DOI] [PubMed] [Google Scholar]
- 23.Gilks, W. P., Abou-Sleiman, P. M., Gandhi, S., Jain, S., Singleton, A., Lees, A. J., Shaw, K., Bhatia, K. P., Bonifati, V., Quinn, N. P., et al. (2005) Lancet 365, 415-416. [DOI] [PubMed] [Google Scholar]
- 24.Deng, H., Le, W., Guo, Y., Hunter, C. B., Xie, W. & Jankovic, J. (2005) Ann. Neurol. 57, 933-934. [DOI] [PubMed] [Google Scholar]
- 25.Paisan-Ruiz, C., Lang, A. E., Kawarai, T., Sato, C., Salehi-Rad, S., Fisman, G. K., Al Khairallah, T., George-Hyslop, P., Singleton, A. & Rogaeva, E. (2005) Neurology 65, 664-665. [DOI] [PubMed] [Google Scholar]
- 26.Bae, B. I., Xu, H., Igarashi, S., Fujimuro, M., Agrawal, N., Taya, Y., Hayward, S. D., Moran, T. H., Montell, C., Ross, C. A., et al. (2005) Neuron 47, 29-41. [DOI] [PubMed] [Google Scholar]
- 27.Chung, K. K., Zhang, Y., Lim, K. L., Tanaka, Y., Huang, H., Gao, J., Ross, C. A., Dawson, V. L. & Dawson, T. M. (2001) Nat. Med. 7, 1144-1150. [DOI] [PubMed] [Google Scholar]
- 28.Engelender, S., Kaminsky, Z., Guo, X., Sharp, A. H., Amaravi, R. K., Kleiderlein, J. J., Margolis, R. L., Troncoso, J. C., Lanahan, A. A., Worley, P. F., et al. (1999) Nat. Genet. 22, 110-114. [DOI] [PubMed] [Google Scholar]
- 29.Smith, W. W., Norton, D. D., Gorospe, M., Jiang, H., Nemoto, S., Holbrook, N. J., Finkel, T. & Kusiak, J. W. (2005) J. Cell Biol. 169, 331-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poirier, M. A., Jiang, H. & Ross, C. A. (2005) Hum. Mol. Genet. 14, 765-774. [DOI] [PubMed] [Google Scholar]
- 31.Smith, W., Margolis, R. L., Li, X., Troncoso, J., Lee, M. K., Dawson, V. L., Dawson, D. M., Iwatsubo, T. & Ross, C. A. (2005) J. Neurosci. 25, 5544-5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung, K. K., Dawson, V. L. & Dawson, T. M. (2001) Trends Neurosci. 24, S7-S14. [DOI] [PubMed] [Google Scholar]
- 33.Chung, K. K., Thomas, B., Li, X., Pletnikova, O., Troncoso, J. C., Marsh, L., Dawson, V. L. & Dawson, T. M. (2004) Science 304, 1328-1331. [DOI] [PubMed] [Google Scholar]
- 34.Moore, D. J., Dawson, V. L. & Dawson, T. M. (2003) Neuromol. Med. 4, 95-108. [DOI] [PubMed] [Google Scholar]
- 35.Ross, O. A. & Farrer, M. J. (2005) Biochem. Soc. Trans. 33, 586-590. [DOI] [PubMed] [Google Scholar]
- 36.Darios, F., Corti, O., Lucking, C. B., Hampe, C., Muriel, M. P., Abbas, N., Gu, W. J., Hirsch, E. C., Rooney, T., Ruberg, M., et al. (2003) Hum. Mol. Genet. 12, 517-526. [DOI] [PubMed] [Google Scholar]
- 37.Kalia, S. K., Lee, S., Smith, P. D., Liu, L., Crocker, S. J., Thorarinsdottir, T. E., Glover, J. R., Fon, E. A., Park, D. S. & Lozano, A. M. (2004) Neuron 44, 931-945. [DOI] [PubMed] [Google Scholar]
- 38.Ko, H. S., von Coelln, R., Sriram, S. R., Kim, S. W., Chung, K. K., Pletnikova, O., Troncoso, J., Johnson, B., Saffary, R., Goh, E. L., et al. (2005) J Neurosci. 25, 7968-7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimura, H., Hattori, N., Kubo, S., Mizuno, Y., Asakawa, S., Minoshima, S., Shimizu, N., Iwai, K., Chiba, T., Tanaka, K., et al. (2000) Nat. Genet. 25, 302-305. [DOI] [PubMed] [Google Scholar]
- 40.Schlossmacher, M. G., Frosch, M. P., Gai, W. P., Medina, M., Sharma, N., Forno, L., Ochiishi, T., Shimura, H., Sharon, R., Hattori, N., et al. (2002) Am. J. Pathol. 160, 1655-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Junn, E., Lee, S. S., Suhr, U. T. & Mouradian, M. M. (2002) J. Biol. Chem. 277, 47870-47877. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka, M., Kim, Y. M., Lee, G., Junn, E., Iwatsubo, T. & Mouradian, M. M. (2004) J. Biol. Chem. 279, 4625-4631. [DOI] [PubMed] [Google Scholar]
- 43.Betarbet, R., Sherer, T. B. & Greenamyre, J. T. (2005) Exp. Neurol 191, Suppl. 1, S17-S27. [DOI] [PubMed] [Google Scholar]
- 44.Ross, C. A. & Pickart, C. (2004) Trends Cell Biol. 14, 703-711. [DOI] [PubMed] [Google Scholar]
- 45.Palacino, J. J., Sagi, D., Goldberg, M. S., Krauss, S., Motz, C., Wacker, M., Klose, J. & Shen, J. (2004) J. Biol. Chem. 279, 18614-18622. [DOI] [PubMed] [Google Scholar]
- 46.Pramstaller, P. P., Schlossmacher, M. G., Jacques, T. S., Scaravilli, F., Eskelson, C., Pepivani, I., Hedrich, K., Adel, S., Gonzales-McNeal, M., Hilker, R., et al. (2005) Ann. Neurol. 58, 411-422. [DOI] [PubMed] [Google Scholar]
- 47.Ross, C. A. & Poirier, M. A. (2004) Nat. Med. 10, Suppl., S10-S17. [DOI] [PubMed] [Google Scholar]
- 48.Ross, C. A. & Poirier, M. A. (2005) Nat. Rev. Mol. Cell Biol. 6, 891-898. [DOI] [PubMed] [Google Scholar]
- 49.Kruger, R. (2004) J. Neurol. 251, Suppl. 6, VI/2-VI/6. [DOI] [PubMed] [Google Scholar]
- 50.Annaert, W., Cupers, P., Saftig, P. & De Strooper, B. (2000) Ann. N.Y. Acad. Sci. 920, 158-164. [DOI] [PubMed] [Google Scholar]
- 51.Selkoe, D. J. (2001) Proc. Natl. Acad. Sci. USA 98, 11039-11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suh, Y. H. & Checler, F. (2002) Pharmacol. Rev. 54, 469-525. [DOI] [PubMed] [Google Scholar]