Improve time and memory of plot_initial_state#664
Improve time and memory of plot_initial_state#664mark-petersen merged 0 commit intoMPAS-Dev:ocean/developfrom
Conversation
TestingSo far, I tested only on |
There was a problem hiding this comment.
This breaks the data set into "chunks" of 32k cells or edges per "chunk". Chunks are distributed across multiple threads (36 on a Grizzly compute node) whenever a computation like min(), max(), xarray.plot.hist(), etc. is performed.
There was a problem hiding this comment.
I always define the index variables right away to be their zero-based python variants.
There was a problem hiding this comment.
This is the key: the xarray version of hist is way better, it seems.
8e4a855 to
8f5310a
Compare
There was a problem hiding this comment.
The xarray version of min and max know to ignore masked (NaN) entries.
There was a problem hiding this comment.
By making these masks into xarray.DataArrays, we can use them to mask other data arrays with where().
There was a problem hiding this comment.
Select time index 0 of the given variable, then mask it by putting NaNs for invalid cells.
8f5310a to
588f322
Compare
| var = ncfile.variables[varName][0, :, :][edgeMask] | ||
| plt.hist(var, bins=100, log=True) | ||
| var = ds[varName].isel(Time=0).where(edgeMask) | ||
| maxRx1Edge = var.max().values |
There was a problem hiding this comment.
compute the max Haney number after masking.
| plt.tight_layout(pad=4.0) | ||
|
|
||
| plt.savefig(args.output_file_name, bbox_inches='tight', pad_inches=0.1) |
There was a problem hiding this comment.
The layout of the figure is a lot better with these settings, taken from MPAS-Analysis climatology map plots.
|
Wow, this is fantastic, and serves as a great example of how to convert a post-processing code to xarray and dask. Thank you, @xylar! I'll test this with upcoming batch of COMPASS changes. |
 mark-petersen
left a comment
mark-petersen
left a comment
There was a problem hiding this comment.
Confirmed that this ran correctly in an EC60to30. Plots look correct. Only took 12s to run! Thanks again!
…3800) Update mpas-source: compass only This PR brings in a new mpas-source submodule with changes only to the ocean core. All of the changes are to the COMPASS testing code that is not used in E3SM, so this PR should have no effect on E3SM. The COMPASS commits are: * MPAS-Dev/MPAS-Model#563 'xylar/ocean/remove_jigsaw_to_mpas' * MPAS-Dev/MPAS-Model#662 'xylar/ocean/fix_broken_compass_tests' * MPAS-Dev/MPAS-Model#664 'xylar/ocean/fix_plot_initial_state' [BFB]
Update mpas-source: compass only This PR brings in a new mpas-source submodule with changes only to the ocean core. All of the changes are to the COMPASS testing code that is not used in E3SM, so this PR should have no effect on E3SM. The COMPASS commits are: * MPAS-Dev/MPAS-Model#563 'xylar/ocean/remove_jigsaw_to_mpas' * MPAS-Dev/MPAS-Model#662 'xylar/ocean/fix_broken_compass_tests' * MPAS-Dev/MPAS-Model#664 'xylar/ocean/fix_plot_initial_state' [BFB]
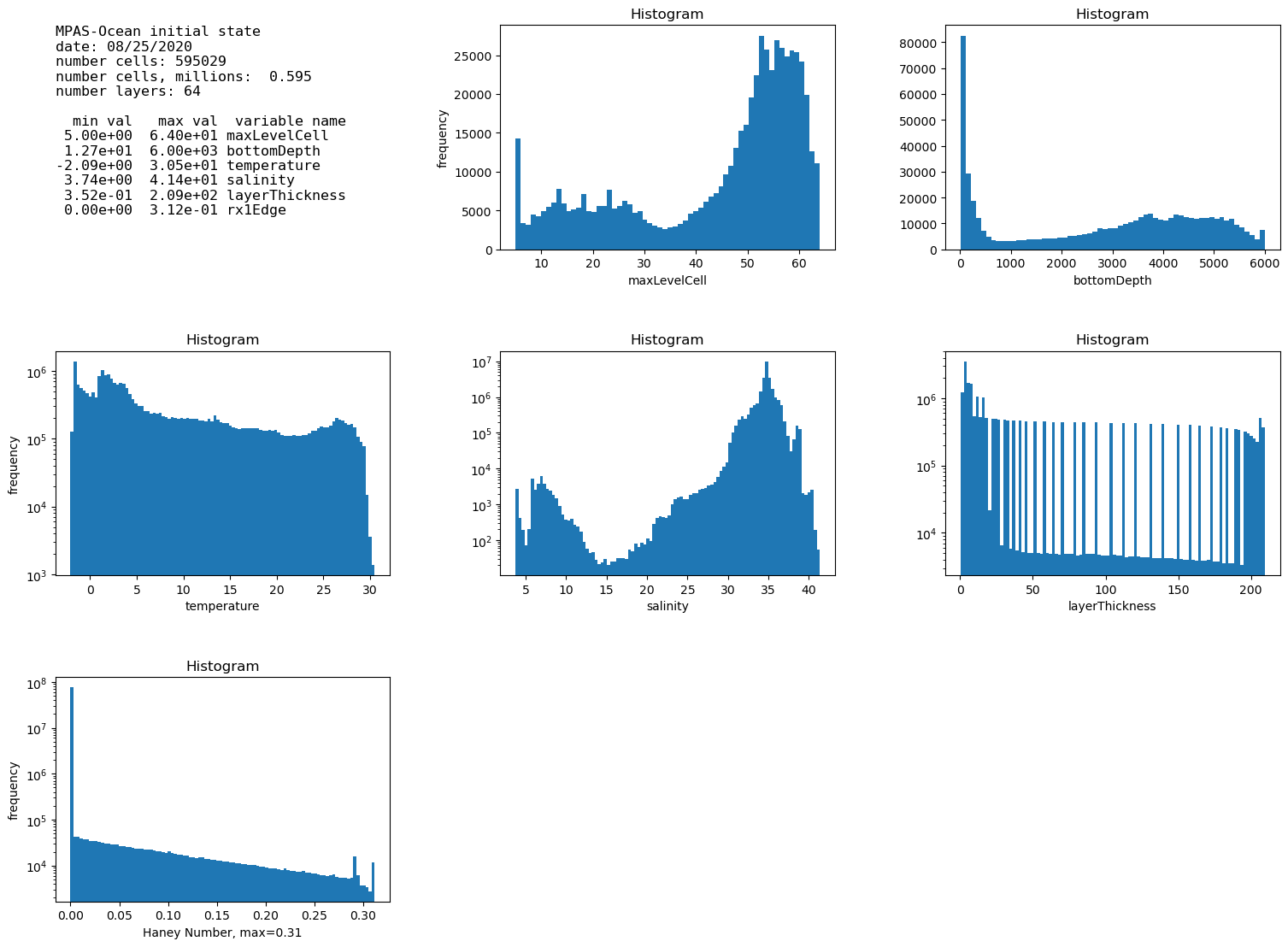
This merge converts plot_initial_state to use
xarraywith chunking to usedaskparallelism. It also usesxarray's version ofhistograms. The plotting of the largest COMPASS mesh (
ARM) goes for timing out or running out of memory even for 3 hour jobs to running in 15 seconds and using about 1% of the memory of a Grizzly compute node.closes #663